Abstract
Gaussian mixture models have many applications in density estimation and data clustering. However, the model does not adapt well to curved and strongly nonlinear data, since many Gaussian components are typically needed to appropriately fit the data that lie around the nonlinear manifold. To solve this problem, the active function cross-entropy clustering (afCEC) method was constructed. In this article, we present an online afCEC algorithm. Thanks to this modification, we obtain a method which is able to remove unnecessary clusters very fast and, consequently, we obtain lower computational complexity. Moreover, we obtain a better minimum (with a lower value of the cost function). The modification allows to process data streams.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Clustering plays a basic role in many parts of data engineering, pattern recognition, and image analysis. Some of the most important clustering methods are based on GMM, which in practice accommodates data with distributions that lie around affine subspaces of lower dimensions obtained by principal component analysis (PCA) [17], see Fig. 1a. However, by the manifold hypothesis, real-world data presented in high-dimensional spaces are likely to be concentrated in the vicinity of nonlinear sub-manifolds of lower dimensionality [4, 30]. The classical approach approximates this manifold by a mixture of Gaussian distributions. Since one non-Gaussian component can be approximated by a mixture of several Gaussians [10, 35, 38], these clusters are, in practice, represented by a combination of Gaussian components. This can be seen as a form of piecewise linear approximation, see Fig. 1a. Cross-entropy clustering (CEC) [34, 36, 37, 40] approach gives similar result.
In [39], authors have constructed the afCEC (active function cross-entropy clustering) algorithm, which allows the clustering of data on sub-manifolds of \({\mathbb {R}}^d\). The motivation comes from the observation that it is often profitable to describe nonlinear data by a smaller number of components with more complicated curved shapes to obtain a better fit of the data, see Fig. 1b. The afCEC method automatically reduces unnecessary clusters and accommodates nonlinear structures.
In this paper, the online version of the afCECFootnote 1 algorithm using Hartigan’s approach is presented. In a case when a new point appears, we are able to update parameters of all clusters without recomputing all variables. Because we have to approximate complicated structures in each step, we have to construct a numerically efficient model. Therefore, we have chosen an approach that allows for the use of an explicit formula in each step.
The algorithm proceeds point by point and determines its optimal cluster assignment. The method only iterates if some cluster has a point closer to some other cluster’s center. Hartigan’s method takes into account the motion of the means resulting from the reassignment—that is, it may reassign a point to another cluster, even if it is already assigned to the closest center.
Thanks to such a modification, the unnecessary clusters are efficiently removed [40], usually in the first three or four iterations. In consequence, one needs smaller number of steps in each iteration to find the local minimum. Moreover, Hartigan’s method finds essentially better minima (with lower cost function value). In Fig. 2, we present the convergence process of Hartigan’s afCEC with the initial number of clusters at \(k=10\), which is reduced to \(k=5\).
The modification also allows processing data streams [33] in which the input is presented as a sequence of items and can be examined in only a few passes (typically just one). These algorithms have limited memory available for them (much less than the input size) and also limited processing time per item. The intrinsic nature of stream data requires the development of algorithms capable of performing fast and incremental processing of data objects. Therefore, Hartigan’s version of afCEC algorithm can be applied in data streams clustering.
The paper is organized as follows. In the next section, related work is presented. In Sect. 3, we introduce afCEC algorithm. In Sect. 4, we present the Hartigan modification of the method. In particular, we discuss how to update parameters online. In the last section, a comparison between our approach and classical algorithms is made.
2 Related works
Clustering is the classical problem of dividing a data \(X \in {\mathbb {R}}^N\) into a collection of disjoint groups \(X_1, \ldots X_k\). Several of the most popular clustering methods are based on the k-means approach [1]. In the context of the algorithm, there were introduced two basic heuristics for minimizing the cost function: Lloyd’s and Hartigan’s. The methods became standards in the general clustering theorem.
The first heuristic for k-means (or general clustering methods) is the Lloyd’s approach: given some initial clustering, we assign points to the closest one [9, 25, 26]. This scheme is intuitive, and empirical support is favorable: the technique generally seems to find a good solution in a small number of iterations. The alternative heuristic was presented by Hartigan [14, 15]: repeatedly pick a point and determine its optimal cluster assignment. The obvious distinction with Lloyd is that the algorithm proceeds point by point. The comparison of the method is presented in [41]. Roughly speaking, in the context of k-means, Hartigan’s approach converges to the minimum faster and generally find better minima of the cost function. On the other hand, Lloyd’s approach is more resistant to outliers.
The basic drawback of k-means algorithm was solved by using density-based techniques, which use expectation maximization (EM) method [27]. The Gaussian mixture model (GMM) is probably the most popular [28, 29]. Thanks to this approach, we can describe clusters by more general shapes like ellipses.
The cross-entropy clustering (CEC) approach [40] joins the clustering advantages of k-means and EM. It turns out that CEC inherits the speed and scalability of k-means, while overcoming the ability of EM to use mixture models. CEC allows an automatic reduction in “unnecessary” clusters, since, contrary to the case of classical k-means and EM, there is a cost of using each cluster. One of the most important properties of CEC, in relation to GMM, is that, similar to k-means, we can use Hartigan’s approach.
Since typically data lie around curved structures (manifold hypotheses), algorithms which can approximate curves or manifolds are important. Principal curves and principal surfaces [16, 18, 22] have been defined as self-consistent smooth curves (or surfaces in \({\mathbb {R}}^2\)) which pass through the middle of a d-dimensional probability distribution or data cloud. They give a summary of the data and also serve as an efficient feature extraction tool.
Another method that attempts to solve the problem of fitting nonlinear manifolds is that of self-organizing maps (SOM) [20], or self-organizing feature maps (SOFM) [19]. These methods are types of artificial neural networks which are trained using unsupervised learning to produce a low-dimensional (typically two-dimensional) discretized representation of the input space of the training samples, called a map.
Kernel methods provide a powerful way of capturing nonlinear relations. One of the most common, kernel PCA (KCPA) [32], is a nonlinear version of principal component analysis (PCA) [17] that gives an explicit low-dimensional space such that the data variance in the feature space is preserved as much as possible.
The above approaches focus on finding only a single complex manifold. In general, they do not focus on the clustering method. Furthermore, it is difficult to use them for dealing with clustering problems. Kernel methods and self-organizing maps can be used as a preprocessing for classical clustering methods. In such a way, spectral clustering methods were constructed [24]. The classical kernel k-means [24] is equivalent to KPCA prior to the conventional k-means algorithm. Spectral clustering is a large family of grouping methods which partition data using eigenvectors of an affinity matrix derived from the data [7, 43,44,45, 48].
The active curve axis Gaussian mixture model (AcaGMM) [47] is an adaptation of the Gaussian mixture model, which uses a nonlinear curved Gaussian probability model in clustering. AcaGMM works well in practice; however, it has major limitations. First of all, the AcaGMM cost function does not necessarily decrease with iterations, which causes problems with the stop condition, see [39]. Since the method uses orthogonal projections and arc lengths, it is very hard to use AcaGMM for more complicated curves in higher-dimensional spaces.
The active function cross-entropy clustering [39] (afCEC) method (see Fig. 1b), which is based on the cross-entropy clustering (CEC) model, solves all the above limitations. The method has a few advantages in relation to AcaGMM: it enables easy adaptation to clustering of complicated datasets along with a predefined family of functions and does not need external methods to determine the number of clusters, as it automatically reduces the number of groups.
In practice, afCEC gives essentially better results than linear models like GMM or CEC, since we obtain a similar level of the Log-likelihood function by using a smaller number of parameters to describe the model. On the other hand, the results are similar to that of AcaGMM when we restrict the data to two dimensions and use the quadratic function as the baseline. For more detailed comparison between the methods, see [39].
All the above approaches do not have Hartigan’s versions. In this article, we present an online afCEC algorithm. In the case of Lloyd’s approach, authors use the regression method for each step. In this paper, we present how to apply Hartigan’s heuristic for minimizing afCEC cost function. Thanks to this modification, we obtain a method which is able to remove unnecessary clusters very fast and, consequently, we obtain a lower computational complexity. Moreover, we obtain a better minimum (with lower value of the cost function).
3 AfCEC algorithm
In this section, we briefly describe AfCEC method (for more information we refer to [39]). At the beginning, we introduce a density distribution which was used in AfCEC method—f-adapted Gaussian density. Let us recall that the standard Gaussian density in \({\mathbb {R}}^d\) is defined by
where \({m}\) denotes the mean, \(\varSigma \) is the covariance matrix, and \( \Vert {\mathrm {v}}\Vert ^2_{\varSigma }={\mathrm {v}}^T \varSigma ^{-1} {\mathrm {v}}\) is the square of the Mahalanobis norm.
In our work, we use a multidimensional Gaussian density in a curvilinear coordinate system which is “spread” along the function \(f :{\mathbb {R}}^{d-1} \rightarrow {\mathbb {R}}\) (f-adapted Gaussian density). We treat one of the variables separately. In such a case, we consider only those \(\varSigma \in {\mathcal {M}}_{d}({\mathbb {R}})\) (where \({\mathcal {M}}_{d}({\mathbb {R}})\) denotes the set of d-dimensional square, symmetrical, and positive define matrices) which have the diagonal block matrix form
where \(\varSigma _{\hat{l}} \in {\mathcal {M}}_{d-1}({\mathbb {R}})\) and \(\varSigma _{l} > 0\). For \({x}= [x_1,\ldots ,x_d]^T \in {\mathbb {R}}^d\) and \(l\in \{1,\ldots ,d\}\), we will use the notation
Now, we will give a mathematically formal definition of the f-adapted Gaussian function.
Definition 1
Let \(f \in {\mathcal {C}}({\mathbb {R}}^{d-1},{\mathbb {R}}) \), \(\varSigma _{\hat{l}} \in {\mathcal {M}}_{d-1}({\mathbb {R}})\), \(\varSigma _{l} > 0\), \({m}=[{m}_{\hat{l}},m_{l}]^T \in {\mathbb {R}}^{d}\) be given. The f-adapted Gaussian density for \(\varSigma _{\hat{l}}\), \(\varSigma _{l}\), \(l\in \{1,\ldots ,d\}\) and \({m}\) is defined as follows:
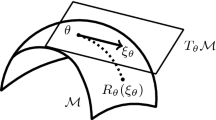
In the basic form of the CEC algorithm [40], we are looking for the optimal Gaussian function in the family of all d-dimensional Gaussian densities \({\mathcal {G}}({\mathbb {R}}^d)\). In the case of AfCEC, we describe each cluster by the f-adapted Gaussian function, see Fig. 3. Consequently, we need to find optimal density in the class of all curved Gaussians. For the given \(f:{\mathbb {R}}^{d-1} \rightarrow {\mathbb {R}}\), we denote the family of all f-adapted Gaussian functions by
In the AfCEC algorithm, we describe clusters by generalized Gaussian distributions from \({\mathcal {A}}_{l}[f]\) where f is in some class of functions (we can use any class of functions for which the regression procedure works) and \(l\in \{1,\ldots ,d\}\). Therefore, we will need one more definition. For the family \({\mathcal {F}}\subset {\mathcal {C}}({\mathbb {R}}^{d-1},{\mathbb {R}})\), we define
In the previous considerations, we assumed that one variable was chosen to be dependent. Since, in the case of the \({\mathcal {F}}\)-adaptive Gaussian density, all computations are applied in the canonical basis, we can verify all possible dependent variable choices. For the family \({\mathcal {F}}\subset {\mathcal {C}}({\mathbb {R}}^{d-1},{\mathbb {R}})\), we define the family of \({\mathcal {F}}\)-adapted Gaussian distributions with all the possible choices of dependent variables by
Since our method is based on the CEC approach, we start with a short introduction to the method (for a more detailed explanation we refer the reader to [40]). To apply CEC, we need to introduce the cost function which we want to minimize. In the case of splitting \(X \subset {\mathbb {R}}^d\) into \(X_1, \ldots , X_k\) so that we code elements of \(X_i\) using a function from the family of all Gaussian densities \({\mathcal {G}}({\mathbb {R}}^d)\), the mean code-length of a randomly chosen element x equals
where \(p_i = \frac{|X_i|}{|X|}\). The formula uses the cross-entropy of a dataset with respect to the family \({\mathcal {G}}({\mathbb {R}}^d)\). In the case of AfCEC, our goal is to calculate an explicit formula for the cost function in the case of f-adapted Gaussian densities.
Optimization Problem 31
Divide the dataset \(X \subset {\mathbb {R}}^d\) into k pairwise disjoint groups \(X_1,\ldots ,X_k\) (\(X= X_1 \cup \ldots \cup X_k\)) such that the cost function
\( \text{ where } p_{i}=\frac{|X_{i}|}{|X|},\) is minimal.
If \({\mathcal {F}}\) is a set of functions which are invariant under the operations \(f \rightarrow a+f\) for any a, we have a following theorem.
Theorem 1
Let \(X \subset {\mathbb {R}}^d\) be a dataset, and let a family of functions \({\mathcal {F}}\subset {\mathcal {C}}( {\mathbb {R}}^{d-1}, {\mathbb {R}}) \) be invariant under the operations \(f \rightarrow a+f\) for \(a \in {\mathbb {R}}\). Let \({\bar{f}}_{l} \in {\mathcal {F}}\) for \(l\in \{1,\ldots ,d\}\) be such that\( {\bar{f}}_{l} = {\text {argmin}}\{ f \in {\mathcal {F}}:|x_{l}-f({x}_{\hat{l}})|^2 \}\). Then,
where \(\varSigma _{\hat{l}} = {\mathrm {cov}}(X_{\hat{l}})\)and \(l \in \{1, \ldots , d \}.\)
We can analyze each cluster separately. For one cluster \(X \subset {\mathbb {R}}^d\), we estimate the parameters of the model in two steps. First, we consider all of the possible choices of dependent variables and calculate functions \(f_{l}\) (corresponding to relations \(x_{l}=f_{l}({x}_{\hat{l}})\)), means \(m_{ l} = {\mathrm {mean}}(X_{l}^{f_{l}})\), \({m}_{\hat{l}} = {\mathrm {mean}}(X_{\hat{l}})\) and covariances \(\varSigma _{\hat{l}} ={\mathrm {cov}}(X_{\hat{l}})\), \( \varSigma _{l} = {\mathrm {cov}}(X_{l}^{f_{l}})\) for \(l\in \{1,\ldots ,d\}\). More precisely, we find \(f_{l}\)-adapted Gaussian distributions \( N([{m}_{\hat{l}},0]^T,\varSigma _{\hat{l}},\varSigma _{l},f_{l}), \) which realize a minimum of cross-entropy \( H^{\times }(X\Vert {\mathcal {A}}_{l}[{\mathcal {F}}]), \) for \(l\in \{1,\ldots ,d\}\). Then we determine the optimal dependent variable \( j = {\text {argmin}}_{l\in \{1,\ldots ,d\}} \{H^{\times }(X\Vert {\mathcal {A}}_{l}[{\mathcal {F}}])\}. \) Consequently, our dataset is represented by the active function, mean, and covariance matrix
where subscript \(j \in \{1,\ldots , d\}\) denotes the dependent variable in the cluster. The above parameters minimize the cost function of one cluster \(H^{\times }(X \Vert {\mathcal {A}}[{\mathcal {F}}])\).
CEC allows an automatic reduction in “unnecessary” clusters, since, contrary to the case of classical k-means and EM, there is a cost of using each cluster. (The step-by-step view of this process is shown in Fig. 2.) There are also several probabilistic approaches which try to estimate the correct number of clusters. For example, [11] uses the generalized distance between Gaussian mixture models with different components number by using the Kullback–Leibler divergence, see [6, 21]. A similar idea is presented by [46] (Competitive Expectation Maximization) which uses the minimum message length criterion provided by [8]. In practice, MDLP can also be directly used in clustering, see [42]. However, most of the above-mentioned methods typically proceed through all the consecutive clusters and do not reduce the number of clusters online during the clustering process.
Classical AfCEC algorithm presented in [39] uses Lloyd’s method. The alternative heuristic was presented by Hartigan [14, 15]: repeatedly pick a point and determine its optimal (from the cost function’s point of view) cluster assignment. Observe that in the crucial step in Hartigan’s approach we compare the cross-entropy after and before the switch, while the switch removes a given point from one cluster and adds it to the other. It means that to apply efficiently the Hartigan approach in clustering it is essential to update parameters (7) when we add a point to the cluster and downdate parameters (7) when we delete a point from group. In the next section, we present how we can update and downdate all parameters of afCEC online.
4 Updating the value of the cost function
Recall that for the particular cluster X our goal is to present how to update and downdate meta-parameters:
Observe that in the crucial step in Hartigan’s approach we compare the cross-entropy after and before the switch. Therefore, it is enough to update/downdate only parameters on which the afCEC cost function
depends. Evaluating the cost function for the dataset \(X \in {\mathbb {R}}^d\) involves computing two quantities: the covariance matrix on \(X_{\hat{d}}\)
and mean squared error (MSE), respectively, on the d-coordinate
along the active axis defined as the mean squared error of the linear least-squares approximation of the data along the active axis.
It should be highlighted that for updating the above parameters we additionally require some other information. In the case of \({\mathrm {cov}}(X_{\hat{d}})\), we need to store the \({\mathrm {mean}}(X_{\hat{d}})\). In such a case, we have a simple formula. Update and downdate procedures are given by the following formulas:
-
(a)
The update procedure:
$$\begin{aligned} \begin{array}{l} {\mathrm {mean}}\left( X_{\hat{d}} \cup \left\{ {x}_{\hat{d}}\right\} \right) = p_{1} {\mathrm {mean}}\left( X_{\hat{d}}\right) +p_{2}{x}_{\hat{d}}, \\ {\mathrm {cov}}\left( X_{\hat{d}} \cup \left\{ {x}_{\hat{d}}\right\} \right) = p_{1}{\mathrm {cov}}\left( X_{\hat{d}}\right) +p_{1}p_{2}\left( {\mathrm {mean}}\left( X_{\hat{d}}\right) -{x}_{\hat{d}}\right) \left( {\mathrm {mean}}\left( X_{\hat{d}}\right) -{x}_{\hat{d}}\right) ^T, \end{array} \end{aligned}$$where \(p_1=\frac{|X|}{|X| + 1}\), \(p_2=\frac{1}{|X|+1}\), and \({x}\notin X\).
-
(b)
The downdate procedure:
$$\begin{aligned} \begin{array}{l} {\mathrm {mean}}\left( X_{\hat{d}} {\setminus} \left\{ {x}_{\hat{d}}\right\} \right) = q_1 {\mathrm {mean}}\left( X_{\hat{d}}\right) -q_2{x}_{\hat{d}}, \\ {\mathrm {cov}}\left( X_{\hat{d}} {\setminus} \left\{ {x}_{\hat{d}}\right\} \right) = q_1{\mathrm {cov}}\left( X_{\hat{d}}\right) -q_1 q_2 \left( {\mathrm {mean}}\left( X_{\hat{d}}\ \right) -{x}_{\hat{d}}\right) \left( {\mathrm {mean}}\left( X_{\hat{d}}\right) -{x}_{\hat{d}}\right) ^T, \end{array} \end{aligned}$$where \(q_1=\frac{|X|}{|X|-1}\), \(q_2=\frac{1}{|X|-1}\), and \({x}\in X\).
In the case of the second quantity \(\frac{1}{|X|}\sum _{{x}\in X}(x_d-f({x}_{\hat{d}}))^2\), we have a more complicated situation. Our goal is to update/downdate the regression function f. There are many types of regression models. In our work, we consider a general one. For a data \(X \subset {\mathbb {R}}^d\), we use the model
where \(f_j\) are linearly independent functions. In the case of \(f_j({x}_{\hat{d}})=x^{j-1}\), we obtain a classical polynomial regression. Therefore, our goal is to find a vector \( \upalpha = \begin{bmatrix} \alpha _1&\ldots&\alpha _m \end{bmatrix}^T, \) which minimizes
Directly from the regression theory, we can calculate the vector \(\upalpha \) as a solution to a system of linear equations.
Theorem 2
Let \(X \in {\mathbb {R}}^d\) be given. Let \(f({x})=\alpha _1 f_1({x}_{\hat{d}}) + \cdots + \alpha _m f_m({x}_{\hat{d}}),\) where \(f_i:{\mathbb {R}}^{d-1}\rightarrow {\mathbb {R}}\) are linearly independent functions for which linear regression is considered. Then, the vector \(\upalpha \) which minimizes the mean squared error
satisfies the following linear equation system:
Similar to the previous situation, we will store additional elements to be able to update/downdate our parameters. For the data X, we denote the matrix from Eq. (14) by \( A_X \) and the vector from Eq. (14) \( {b}_X. \) Consequently, Eq. (14) can be rewritten in the following form
The main idea is to update/downdate parameters \(A_X\) and \({b}_X\) and solve the linear equation in each iteration to determine the updated \(\upalpha \).
Theorem 3
Let \(X \in {\mathbb {R}}^d\) be given. Let \(f({x}_{\hat{d}})=\alpha _1 f_1({x}_{\hat{d}}) + \cdots + \alpha _m f_m({x}_{\hat{d}})\), where \(f_i:{\mathbb {R}}^{d-1}\rightarrow {\mathbb {R}}\) are linearly independent functions for which linear regression is considered. For \({x}\), update (\({x}\notin X)\) and downdate (\({x}\in X)\) procedures are given by the following formulas:
-
(a)
The update procedure:
$$\begin{aligned} {\mathrm {A}}_{X \cup \{{x}\}}={\mathrm {A}}_X + \begin{bmatrix} f_1\left( {x}_{\hat{d}}\right) \\ \vdots \\ f_m\left( {x}_{\hat{d}}\right) \end{bmatrix} \begin{bmatrix} f_1\left( {x}_{\hat{d}}\right) \\ \vdots \\ f_m\left( {x}_{\hat{d}}\right) \end{bmatrix}^T, \end{aligned}$$(16)$$\begin{aligned} {b}_{X \cup \{{x}\}}={b}_X + \begin{bmatrix} f_1\left( {x}_{\hat{d}}\right) \\ \vdots \\ f_m\left( {x}_{\hat{d}}\right) \end{bmatrix} x_d. \end{aligned}$$(17) -
(b)
The downdate procedure:
$$\begin{aligned} {\mathrm {A}}_{X \setminus \{{x}\}}={\mathrm {A}}_X - \begin{bmatrix} f_1\left( {x}_{\hat{d}}\right) \\ \vdots \\ f_m\left( {x}_{\hat{d}}\right) \end{bmatrix} \begin{bmatrix} f_1\left( {x}_{\hat{d}}\right) \\ \vdots \\ f_m\left( {x}_{\hat{d}}\right) \end{bmatrix}^T, \end{aligned}$$(18)$$\begin{aligned} {b}_{X \setminus \{{x}\}}={b}_X - \begin{bmatrix} f_1\left( {x}_{\hat{d}}\right) \\ \vdots \\ f_m\left( {x}_{\hat{d}}\right) \end{bmatrix} x_d. \end{aligned}$$(19)
Proof
Let X and \({\bar{{x}}}\) be given. A simple corollary from Theorem 2 is that the vector \(\upalpha \) which minimizes the mean squared error satisfies the following linear equation system:
Now we add/remove \({\bar{{x}}}\) from X. Therefore, by Theorem 2 for \({X \cup \{{\bar{{x}}} \}}\) or \({X \setminus \{{\bar{{x}}}\}}\), respectively, we obtain
\(\square \)
Thanks to Theorem 3, we can update parameters \(A_X\) and \({b}_X\). Then we solve the system of linear equations \( A_X \upalpha ={b}_{X}. \) Therefore, we obtain \(\upalpha \) for \(X\cup \{{x}\}\) or \(X\setminus \{{x}\}\), respectively. In the last step, we can update and downdate the mean squared error (MSE), respectively, on the d-coordinate by using a new value of \(A,{b}, \upalpha \).
Theorem 4
Let\(X \in {\mathbb {R}}^d\)be given. Let\(f({x}_{\hat{d}})=\alpha _1 f_1({x}_{\hat{d}}) + \cdots + \alpha _m f_m({x}_{\hat{d}})\), where\(f_i:{\mathbb {R}}^{d-1}\rightarrow {\mathbb {R}}\)are linearly independent functions for which linear regression is considered. Then
where
Proof
We have
\(\square \)
This can be always done, provided that the matrix \(\varSigma _X\) or \(\varSigma _{X \setminus \{{x}\}}\) (depending on whether we add or remove the point \({x}\) from the dataset X) is non-singular. Having the values of \(\{\alpha _1,\ldots ,\alpha _k\}\), one can immediately obtain the desired value.
5 Algorithm
In this section, we present our algorithm. The aim of the Hartigan’s method is to find a partition \(X_1,\ldots ,X_n\) of X, for which the cost function (4) is as close as possible to the minimum, by subsequently reassigning membership of elements from X.
To explain Hartigan’s approach more precisely, we need the notion of a group membership function \( {\mathrm{gr}} : \{1,\ldots ,n\} \rightarrow \{ 0,\ldots ,k\}, \) which describes the membership of the ith element, where 0 value is a special symbol which denotes that \({x}_i\) is as yet unassigned. In other words, if \({\mathrm{gr}}(i) = l > 0\), then \({x}_i\) is a part of the lth group, and if \({\mathrm{gr}}(i) = 0\) then \({x}_i\) is unassigned.
In Algorithm 1, we present a pseudo-code of the method. The algorithm starts from an initial clustering, which can be obtained randomly or with the use of the k-means++. In our case, we assume that we have an initial clustering given by \({\mathrm {cl}}\). (The number of clusters is given by k.) At the beginning, the algorithm calculates the initial values of parameters which describe each cluster.

We want to find such \({\mathrm {gr}}:\{1,\ldots ,n\} \rightarrow \{1,\ldots ,k\}\) (thus all elements of X are assigned) that \( E(X_1, \ldots , X_k ;\,\,{\mathcal {A}}[{\mathcal {F}}] ) \) is minimal. The basic idea of Hartigan is relatively simple—we repeatedly go over all elements of the partition \(X=(x_i)_{i=1}^n\) and apply the following steps:
-
If the chosen set \(x_i\) is unassigned, assign it to the first non-empty group;
-
Reassign \(x_i\) to that group for which the decrease in cross-entropy is maximal;
-
Check if no group needs to be removed/unassigned, if this is the case unassign its all elements;
until no group membership has been changed.
To implement Hartigan’s approach, we still have to add a condition regarding when to unassign a given group. For example, in the case of AfCEC clustering in \({\mathbb {R}}^d\), to avoid overfitting we cannot consider clusters which contain less then \(d + 1\) points. In practice while applying Hartigan’s approach on discrete data, we usually remove clusters which contain less then five percent of all dataset.
Observe that in the crucial step in Hartigan’s approach, we compare the cross-entropy after and before the switch, while the switch removes a given point from one cluster and adds it to the other. It means that to apply the Hartigan approach efficiently in clustering, it is essential to update/downdate parameters when we add/delete a point from a group by using formulas from Sect. 4.
6 Experiments
In this section, we present a comparison of the Hartigan version of afCEC with density-based methods: GMM, CEC, and Lloyd’s afCEC. It is difficult to compare methods, which use different number of parameters to approximate data. In general, if we use a more complex model, we can fit the data better. Therefore, we use indexes which measure level of fitting and use penalty for using more complicated models.
Hence, there is a trade-off: the better fit, created by making a model more complex by requiring more parameters, must be considered in light of the penalty imposed by adding more parameters.
To compare the results, we use the standard Akaike information criterion (AIC):
and Bayesian information criterion (BIC):
where k is the number of parameters in the model, n is the number of points, and LL is a maximized value of the log-likelihood function.
Let’s analyze the two components of the AIC. The first component, \(-\, 2\)LL, is the value of the likelihood function, which is the probability of obtaining the data given the candidate model. It measures how well the data are fitted by the model. Since the likelihood function’s value is multiplied by \(-\,2\), ignoring the second component, the model with the minimum AIC is the one with the highest value of the likelihood function.
However, to this first component we add an adjustment based on the number of estimated parameters. The more parameters, the greater the amount added to the first component, increasing the value for the AIC and penalizing the model. Hence, there is a trade-off: the better fit, created by making a model more complex by requiring more parameters, must be considered in light of the penalty imposed by adding more parameters. This is why the second component of the AIC is thought of in terms of a penalty.
The Bayesian information criterion (BIC) is another model selection criterion based on information theory but set within a Bayesian context. The difference between the BIC and the AIC is the greater penalty imposed for the number of parameters by the former than the latter.
Consequently, we need a number of parameters which are used in each model. In the case of \({\mathbb {R}}^2\), afCEC uses two scalars for the mean, three scalars for the covariance matrix, and three scalars for the parabola. It should be emphasized that in afCEC, we need to remember which coordinate is the dependent one. This parameter is discrete, so we do not consider it in our investigation.
6.1 The computational times
We compared the computational times between Hartigan version of afCEC and alternative methods: CEC implemented in R package CEC [37, 40] and GMM from R package Rmixmod [23]. We varied the number of dataset instances and the dimension of the data, see Fig. 4. For this purpose, a simple ball-like set was considered.
Comparison of computational efficiency between afCEC, CEC, and GMM. a Comparison of computational efficiency between afCEC, CEC, and GMM in the case of data with different dimensions. b Comparison of computational efficiency between afCEC, CEC, and GMM in the case of data with different number of elements
One can observe that in the case of higher dimensions both afCEC methods give slightly worse results since the regression function must be fitted with respect to all possible dependent variables. It should be highlighted that the application of afCEC method in high-dimensional spaces is rather limited. CEC and GMM methods give comparable results for large datasets.
In the case of data with an increasing number of elements, we can observe that afCEC method gives comparable results to the GMM approach. The method can be applied even to reasonably large datasets. We can observe that Lloyd’s approach gives slightly better results than Hartigan’s algorithm, since we do not have to update parameters in each step. But the use of an online version of the method allows to obtain a better minimum of the cost function and consequently, better clustering, see Tables 1 and 2.
6.2 2D dataset
Let us start with a synthetic dataset. At first, we report the results of afCEC, GMM, CEC in the case of simple 2D sets, see the first two examples in Table 1. As we see, for similar values of the log-likelihood function, we have to use less clusters for afCEC than in GMM and CEC. Moreover, Hartigan’s approach gives better results than Lloyd’s method.
Chinese characters consist of straight-line strokes (horizontal, vertical) and curved strokes (slash, backslash and many types of hooks). GMM has already been employed for analyzing the structure of Chinese characters and has achieved commendable performance [46]. However, some lines extracted by GMM may be too short, and it is quite difficult to join these short lines to form semantic strokes due to the ambiguity of joining them together. This problem becomes more serious when analyzing handwritten characters by GMM, and this was the motivation to use afCEC to represent Chinese characters, see Fig. 5.
In the case of the characters

(cat) for similar values of the log-likelihood function, we have to use 25 clusters for Hartigan afCEC and 35 for GMM and CEC. On the other hand, for simpler characters

(dog),

(father) , we have to use 6 clusters for Hartigan afCEC and 10 for GMM and CEC, see Table 1.
In general, afCEC method usually obtained clustering with the largest value of MLE function. The cost of using additional parameters is small and, consequently, afCEC gives a better clustering in respect to AIC and BIC criteria, see Fig. 6a.
6.3 3D scans of objects
In this subsection, we present how our method works in the case of segmentation of 3D objects. Similarly as before, we report the results of afCEC, GMM, CEC, see Table 2. We show how the log-likelihood, BIC, and AIC functions change when the number of clusters increases. As we can see, for similar values of the log-likelihood function, we have to use less clusters for afCEC than for GMM and CEC. Moreover, we also obtain a better value of BIC and AIC, see the last three examples in Table 2.
The effect of afCEC on 3D objects [2, 3] is shown in Fig. 7. Since afCEC is able to cluster data on sub-manifolds of \({\mathbb {R}}^d\), it is able to fit strongly nonlinear structures of 3D scans of objects. Moreover, afCEC method automatically reduces unnecessary clusters which allows to reduce too small components.
Similar to the previous experiments, afCEC method usually obtained clusterings with the largest value of MLE function. The cost of using additional parameters is small, and consequently, afCEC gives better clusterings with respect to AIC and BIC criteria, see Fig. 6b.
6.4 Comparison with non-density-based methods
Now we present a comparison between afCEC and classical approaches dedicated to clustering of nonlinear datasets: kkmeans [24] and spectral clustering [31] (see Fig. 8). We also use recent modification of the classical method dedicated to nonlinear data STSC [45], SMMC [43], and SSC [7, 44, 48]. In this subsection, we compare algorithms with respect to Rand and Jaccard indexes, see Fig. 9.
Kernel methods can be used as a preprocessing for classical clustering methods. In such a way, spectral clustering methods were constructed [5, 24, 31]. The classical kernel k-means [24] is equivalent to KPCA prior to the conventional k-means algorithm. Most of kernel methods consist of two steps: an embedding into a feature space and a classical clustering method used on the data transformed to feature space. Therefore, spectral methods are typically time-consuming and use large number of parameters.
In the case of datasets of dimension higher than three and afCEC approach, due to computational profitability, we used a smaller class of quadratic polynomials of the type
instead of the class of all quadratic polynomials
This allows us to fit less parameters in each step, which results in a smaller risk of overfitting and helps to effectively cluster higher-dimensional data.
In the case of non-density-based method, we use classical Rand and Jaccard indexes. As we see in Fig. 9 afCEC method gives similar results to other approaches and the Hartigan modification allows to obtain better score.
Comparison between classical data stream clustering algorithms and afCEC over the evolving data stream. a Dataset DSD_Benchmark(1) from R package stream(reproduced with permission from [12]). b Curve-type set. c Rand index for the four classical data stream clustering algorithms and afCEC over the evolving data stream in the case of image from Fig. 10a. d Rand index for the four classical data stream clustering algorithms and afCEC over the evolving data stream in the case of image from Fig. 10b
6.5 Data streams
Typical statistical and data mining methods, including clustering, work with “static” datasets, meaning that the complete dataset is available as a whole to perform all necessary computations. However, in recent years more and more applications need to work with data which is not static but is the result of a continuous data generating process which is likely to evolve over time. This type of data is called a data stream, and dealing with data streams has become an increasingly important area of research.
The characteristic of continually arriving data points introduces an important property of data streams, which also poses the great challenge: the size of a data stream is potentially unbounded. This leads to the following requirements for data stream processing algorithms:
-
Bounded storage The algorithm can only store a very limited amount of data to summarize the data stream;
-
Single pass The incoming data points cannot be permanently stored and need to be processed at once in the arriving order;
-
Real-time The algorithm has to process data points on average at least as fast as the data is arriving;
-
Concept drift The algorithm has to be able to deal with a data generating process which evolves over time (e.g., distributions change or a new structure in the data appears).
In this section, we present a possible application of afCEC method to stream data. We will use the R package stream [12]. In our experiments, we use DSD_Benchmark(1) (see Fig. 10a) form stream, which contains two clusters moving in a two-dimensional space. One moves from the top left to the bottom right and the other one moves from the bottom left to the top right. Both clusters overlap when they meet exactly in the center of the data space. Figure 10a shows plots where clusters move over time. Arrows are added to highlight the direction of cluster movement.
Figure 10c shows the Rand index for the four data stream clustering algorithms and afCEC method over the evolving data stream. All algorithms show that separating the two clusters is impossible around position 3000 when the two clusters overlap. It should be highlighted that afCEC method has a problem with reconstructing the model after the merge. The number of clusters was reduced and cannot be reconstructed. It is possible to add a split merge strategy [13] which would allow to refit afCEC model. The second possible strategy is to add an additional dimension with time components. Since afCEC is an affine invariant it does not change clustering structures and allows to keep two clusters without reduction.
In general, afCEC works in the case when a dataset contains curve-type structures. In the second example, we present how the methods work on such data. Similar to the previous examples, we consider two clusters (two curve-type clusters), where the first moves from top left to bottom right, and the other one moves from bottom left to top right. Figure 10b shows plots where the clusters move over time. Arrows are added to highlight the direction of cluster movement. Figure 10d shows the Rand index. As we see, AfCEC is able to almost perfectly recover the original clustering .
7 Conclusions
In this paper, the Hartigan approach to afCEC method for clustering curved data, which uses generalized Gaussian distributions in curvilinear coordinate systems, was presented. The afCEC method has a strong theoretical background. Moreover, afCEC can be used as a density estimation model. Since afCEC is an implementation of the cross-entropy clustering approach, the method reduces unnecessary clusters online.
In practice, the algorithm gives essentially better results than linear models, like GMM or CEC and the classical Lloyd’s approach to afCEC, since we obtain a similar level of the Log-likelihood function by using a smaller number of parameters to describe the model. Moreover, the online version of afCEC method can be use in the case of stream data.
In the future, we want to update our algorithm to allow the use of closed curves. Thanks to such a modification, we will able either to find more complicated shapes in data or to better adapt to the data structure.
Notes
The Hartigan’s as well as classical Lloyd’s approaches are included in R package afCEC https://cran.r-project.org/web/packages/afCEC/index.html.
References
Bock HH (2007) Clustering methods: a history of K-Means algorithms. In: Bock HH (ed) Selected contributions in data analysis and classification. Springer, Berlin, pp 161–172
Bronstein AM, Bronstein MM, Kimmel R (2006) Efficient computation of isometry-invariant distances between surfaces. SIAM J Sci Comput 28(5):1812–1836
Bronstein AM, Bronstein MM, Kimmel R (2008) Numerical geometry of non-rigid shapes. Springer, Berlin
Cayton L (2005) Algorithms for manifold learning. Univ Calif San Diego Tech Rep 12:1–17
Chi SC, Yang CC (2006) Integration of ant colony SOM and k-means for clustering analysis. In: International conference on knowledge-based and intelligent information and engineering systems, Springer, Berlin, pp 1–8
Cover TM, Thomas JA (2012) Elements of information theory. Wiley, Hoboken
Elhamifar E, Vidal R (2013) Sparse subspace clustering: algorithm, theory, and applications. IEEE Trans Pattern Anal Mach Intell 35(11):2765–2781
Figueiredo MAT, Jain AK (2002) Unsupervised learning of finite mixture models. IEEE Trans Pattern Anal Mach Intell 24(3):381–396
Forgy EW (1965) Cluster analysis of multivariate data: efficiency versus interpretability of classifications. Biometrics 21:768–769
Fraley C, Raftery AE (1998) How many clusters? Which clustering method? Answers via model-based cluster analysis. Comput J 41(8):578–588
Goldberger J, Roweis ST (2004) Hierarchical clustering of a mixture model. In: Proceedings of advances in neural information processing systems, pp 505–512
Hahsler M, Bolanos M, Forrest J (2017) Introduction to stream: an extensible framework for data stream clustering research with R. J Stat Softw 76(14):1–50
Hajto K, Kamieniecki K, Misztal K, Spurek P (2017) Split-and-merge tweak in cross entropy clustering. In: IFIP international conference on computer information systems and industrial management, Springer, Berlin, pp 193–204
Hartigan JA (1975) Clustering algorithms. Wiley, New York
Hartigan JA, Wong MA (1979) Algorithm as 136: a k-means clustering algorithm. Appl Stat 28:100–108
Hastie T, Stuetzle W (1989) Principal curves. J Am Stat Assoc 84(406):502–516
Jolliffe I (2002) Principal component analysis. Encycl Stat Behav Sci 30:487
Kegl BA (1999) Principal curves: learning, design, and applications. Ph.D. Thesis, Citeseer
Kohonen T (1989) Self-organizing feature maps. Springer, Berlin
Kohonen T (2001) Self-organizing maps, vol 30. Springer, Berlin
Kullback S (1997) Information theory and statistics. Dover Pubns, Mineola
LeBlanc M, Tibshirani R (1994) Adaptive principal surfaces. J Am Stat Assoc 89(425):53–64
Lebret R, Iovleff S, Langrognet F, Biernacki C, Celeux G, Govaert G (2015) Rmixmod: the R package of the model-based unsupervised, supervised and semi-supervised classification mixmod library. J Stat Softw 67:241–270
Li J, Li X, Tao D (2008) KPCA for semantic object extraction in images. Pattern Recognit 41(10):3244–3250
Lloyd S (1982) Least squares quantization in PCM. IEEE Trans Inf Theor 28(2):129–137
MacQueen J et al (1967) Some methods for classification and analysis of multivariate observations. In: Proceedings of the fifth Berkeley symposium on mathematical statistics and probability, vol 1. Oakland, pp 281–297
McLachlan G, Krishnan T (1997) The EM algorithm and extensions, vol 274. Wiley, Hoboken
McLachlan G, Krishnan T (2007) The EM algorithm and extensions, vol 382. Wiley, Hoboken
McLachlan G, Peel D (2004) Finite mixture models. Wiley, Hoboken
Narayanan H, Mitter S (2010) Sample complexity of testing the manifold hypothesis. In: Proceedings of advances in neural information processing systems, pp 1786–1794
Ng AY, Jordan MI, Weiss Y et al (2002) On spectral clustering: analysis and an algorithm. Adv Neural Inf Process Syst 2:849–856
Schölkopf B, Smola A, Müller KR (1998) Nonlinear component analysis as a kernel eigenvalue problem. Neural Comput 10(5):1299–1319
Silva JA, Faria ER, Barros RC, Hruschka ER, de Carvalho AC, Gama J (2013) Data stream clustering: a survey. ACM Comput Surv (CSUR) 46(1):13
Śmieja M, Geiger BC (2017) Semi-supervised cross-entropy clustering with information bottleneck constraint. Inf Sci 421:254–271
Śmieja M, Wiercioch M (2016) Constrained clustering with a complex cluster structure. Adv Data Anal Classif 11:1–26
Spurek P (2017) General split gaussian cross-entropy clustering. Expert Syst Appl 68:58–68
Spurek P, Kamieniecki K, Tabor J, Misztal K, Śmieja M (2017) R package CEC. Neurocomputing 237:410–413
Spurek P, Pałk, W (2016) Clustering of gaussian distributions. In: 2016 IEEE international joint conference on neural networks (IJCNN), pp 3346–3353
Spurek P, Tabor J, Byrski K (2017) Active function cross-entropy clustering. Expert Syst Appl 72:49–66
Tabor J, Spurek P (2014) Cross-entropy clustering. Pattern Recognit 47(9):3046–3059
Telgarsky M, Vattani A (2010) Hartigan’s method: k-means clustering without voronoi. In: International conference on artificial intelligence and statistics, pp 820–827
Wallace RS, Kanade T (1990) Finding natural clusters having minimum description length. In: 10th IEEE international conference on proceedings of pattern recognition, 1990, vol 1. pp 438–442
Wang Y, Jiang Y, Wu Y, Zhou ZH (2011) Spectral clustering on multiple manifolds. IEEE Trans Neural Netw 22(7):1149–1161
Yan Q, Ding Y, Xia Y, Chong Y, Zheng C (2017) Class-probability propagation of supervised information based on sparse subspace clustering for hyperspectral images. Remote Sens 9(10):1017
Zelnik-Manor L, Perona P (2005) Self-tuning spectral clustering. In: Advances in neural information processing systems, pp 1601–1608
Zhang B, Zhang C, Yi X (2004) Competitive em algorithm for finite mixture models. Pattern Recognit 37(1):131–144
Zhang B, Zhang C, Yi X (2005) Active curve axis gaussian mixture models. Pattern Recognit 38(12):2351–2362
Zhang H, Zhai H, Zhang L, Li P (2016) Spectral-spatial sparse subspace clustering for hyperspectral remote sensing images. IEEE Trans Geosci Remote Sens 54(6):3672–3684
Acknowledgements
The work of P. Spurek was supported by the National Centre of Science (Poland) Grant No. 2015/19/D/ST6/01472. The work of K. Byrski was supported by the National Centre of Science (Poland) Grant No. 2015/19/D/ST6/01472. The work of J. Tabor was supported by the National Centre of Science (Poland) Grant No. 2017/25/B/ST6/01271.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Spurek, P., Byrski, K. & Tabor, J. Online updating of active function cross-entropy clustering. Pattern Anal Applic 22, 1409–1425 (2019). https://doi.org/10.1007/s10044-018-0701-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10044-018-0701-8