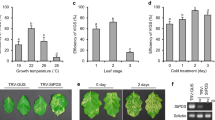
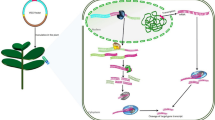
Abstract
Plants have developed defense mechanisms against viruses by using an RNA silencing-based process, which has many common features with the endogenous RNA silencing pathway used for regulating the level of transcripts derived from developmental genes. In the virus-induced gene silencing (VIGS) method, it is possible to take advantage of this mechanism by inserting a plant nucleic fragment within the viral genome to knock down the corresponding gene. This tool has been used in many species as a fast and easy reverse genetics technique in order to gain information on the role of genes with poorly understood functions. Here we describe in detail two Agrobacterium-mediated infection protocols in flax, based on a whole plant vacuum infiltration and a leaf syringe infiltration that systemically impact the transcript levels in the stem.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Baulcombe DC (1999) Fast forward genetics based on virus-induced gene silencing. Curr Opin Plant Biol 2:109–113. https://doi.org/10.1016/S1369-5266(99)80022-3
Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP (2004) Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 39:734–746. https://doi.org/10.1111/j.1365-313X.2004.02158.x
Huis R, Morreel K, Fliniaux O et al (2012) Natural hypolignification is associated with extensive oligolignol accumulation in flax stems. Plant Physiol 158:1893–1915. https://doi.org/10.1104/pp.111.192328
Day A, Ruel K, Neutelings G et al (2005) Lignification in the flax stem: evidence for an unusual lignin in bast fibers. Planta 222:234–245. https://doi.org/10.1007/s00425-005-1537-1
Caillot S, Rosiau E, Laplace C, Thomasset B (2009) Influence of light intensity and selection scheme on regeneration time of transgenic flax plants. Plant Cell Rep 28:359–371. https://doi.org/10.1007/s00299-008-0638-2
Ratcliff F, Martin-Hernandez AM, Baulcombe DC (2001) Technical Advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25:237–245. https://doi.org/10.1046/j.0960-7412.2000.00942.x
Brunt AA (Alan A), International CAB (1996) Viruses of plants: descriptions and lists from the VIDE database/edited by Alan Brunt … [et al.]. CAB International, Wallingford, Oxon, UK
Qin G, Gu H, Ma L et al (2007) Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res 17:471–482. https://doi.org/10.1038/cr.2007.40
Yamagishi M, Masuta C, Suzuki M, Netsu O (2015) Peanut stunt virus-induced gene silencing in white lupin (Lupinus albus). Plant Biotechnol 32:181–191. https://doi.org/10.5511/plantbiotechnology.15.0521a
Wang Z, Hobson N, Galindo L et al (2012) The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J 72:461–473. https://doi.org/10.1111/j.1365-313X.2012.05093.x
Chantreau M, Chabbert B, Billiard S et al (2015) Functional analyses of cellulose synthase genes in flax (Linum usitatissimum) by virus-induced gene silencing. Plant Biotechnol J 13:1312–1324. https://doi.org/10.1111/pbi.12350
Galindo-González L, Deyholos MK (2016) RNA-seq transcriptome response of flax (Linum usitatissimum L.) to the pathogenic fungus Fusarium oxysporum f. sp. lini. Front Plant Sci 7:1–22. https://doi.org/10.3389/fpls.2016.01766
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Chantreau, M., Neutelings, G. (2020). Virus-Induced Gene Silencing of Cell Wall Genes in Flax (Linum usitatissimum). In: Courdavault, V., Besseau, S. (eds) Virus-Induced Gene Silencing in Plants. Methods in Molecular Biology, vol 2172. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0751-0_6
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0751-0_6
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0750-3
Online ISBN: 978-1-0716-0751-0
eBook Packages: Springer Protocols