Abstract
We propose a novel, multi-task, fully convolutional network (FCN) architecture for automatic segmentation of brain tumor. This network extracts multi-level contextual information by concatenating hierarchical feature representations extracted from multimodal MR images along with their symmetric-difference images. It achieves improved segmentation performance by incorporating boundary information directly into the loss function. The proposed method was evaluated on the BRATS13 and BRATS15 datasets and compared with competing methods on the BRATS13 testing set. Segmented tumor boundaries obtained were better than those obtained by single-task FCN and by FCN with CRF. The method is among the most accurate available and has relatively low computational cost at test time.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
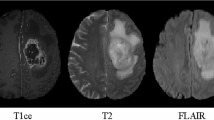
We address the problem of automatic segmentation of brain tumors. Specifically, we present and evaluate a method for tumor segmentation in multimodal MRI of high-grade (HG) glioma patients. Reliable automatic segmentation would be of considerable value for diagnosis, treatment planning and follow-up [1]. The problem is made challenging by diversity of tumor size, shape, location and appearance. Figure 1 shows an HG tumor with expert delineation of tumor structures: edema (green), necrosis (red), non-enhancing (blue) and enhancing (yellow). The latter three form the tumor core.
A common approach is to classify voxels based on hand-crafted features and a conditional random field (CRF) incorporating label smoothness terms [1, 2]. Alternatively, deep convolutional neural networks (CNNs) automatically learn high-level discriminative feature representations. When CNNs were applied to MRI brain tumor segmentation they achieved state-of-the-art results [3,4,5]. Specifically, Pereira et al. [3] trained a 2D CNN as a sliding window classifier, Havaei et al. [4] used 2D CNN on larger patches in a cascade to capture both local and global contextual information, and Kamnitsas et al. [5] trained a 3D CNN on 3D patches and considered global contextual features via downsampling, followed by a fully-connected CRF [6]. All these methods operated at the patch level. Fully convolutional networks (FCNs) recently achieved promising results for natural image segmentation [11, 12] as well as medical image segmentation [13,14,15]. In FCNs, fully connected layers are replaced by convolutional kernels; upsampling or deconvolutional layers are used to transform back to the original spatial size at the network output. FCNs are trained end-to-end (image-to-segmentation map) and have computational efficiency advantages over CNN patch classifiers.
Here we adopt a multi-task learning framework based on FCNs. Our model is a variant of [14,15,16]. Instead of using 3 auxiliary classifiers for each upsampling path for regularization as in [14], we extract multi-level contextual information by concatenating features from each upsampling path before the classification layer. This also differs from [16] which performed only one upsampling in the region task. Instead of either applying threshold-based fusion [15] or a deep fusion stage based on a pooling-upsampling FCN [16] to help separate glands, we designed a simple combination stage consisting of three convolutional layers without pooling, aiming at improving tumour boundary segmentation accuracy. Moreover, our network enables multi-task joint training while [16] has to train different tasks separately, followed by a fine-tuning of the entire network.
Our main contributions are: (1) we are first to apply a multi-task FCN framework to multimodal brain tumor (and substructure) segmentation; (2) we propose a boundary-aware FCN that jointly learns to predict tumor regions and tumor boundary without the need for post-processing, an advantage compared to the prevailing CNN+CRF framework [1]; (3) we demonstrate that the proposed network improves tumor boundary accuracy (with statistical significance); (4) we compare directly using BRATS data; our method ranks top on BRATS13 test data while having good computational efficiency.
2 Variant of FCN
Our FCN variant includes a down-sampling path and three up-sampling paths. The down-sampling path consists of three convolutional blocks separated by max pooling (yellow arrows in Fig. 2). Each block includes 2–3 convolutional layers as in the VGG-16 network [7]. This down-sampling path extracts features ranging from small-scale low-level texture to larger-scale, higher-level features. For the three up-sampling paths, the FCN variant first up-samples feature maps from the last convolutional layer of each convolutional block such that each up-sampled feature map (purple rectangles in Fig. 2) has the same spatial size as the input to the FCN. Then one convolutional layer is added to each up-sampling path to encode features at different scales. The output feature maps of the convolutional layer along the three up-sampling paths are concatenated before being fed to the final classification layer. We used ReLU activation functions and batch normalization. This FCN variant has been experimentally evaluated in a separate study [8].
Variant of FCN. Images and symmetry maps are concatenated as the input to the net [8]. Colored rectangles represent feature maps with numbers nearby being the number of feature maps. Best viewed in color.
3 Boundary-Aware FCN
The above FCN can already produce good probability maps of tumor tissues. However, it remains a challenge to precisely segment boundaries due to ambiguity in discriminating pixels around boundaries. This ambiguity arises partly because convolution operators even at the first convolutional layer lead to similar values in feature maps for those neighboring voxels around tumor boundaries. Accurate tumor boundaries are important for treatment planning and surgical guaidance. To this end, we propose a deep multi-task network.
The structure of the proposed boundary-aware FCN is illustrated in Fig. 3. Instead of treating the segmentation task as a single pixel-wise classification problem, we formulate it within a multi-task learning framework. Two of the above FCN variants with shared down-sampling path and two different up-sampling branches are applied for two separate tasks, one for tumor tissue classification (‘region task’ in Fig. 3) and the other for tumor boundary classification (‘boundary task’ in Fig. 3). Then, the outputs (i.e., probability maps) from the two branches are concatenated and fed to a block of two convolutional layers followed by the final softmax classification layer (‘combination stage’ in Fig. 3). This combination stage is trained with the same objective as the ‘region task’. The combination stage considers both tissue and boundary information estimated from the ‘region task’ and the ‘boundary task’. The ‘region task’ and the ‘combination stage’ task are each a 5-class classification task whereas the ‘boundary task’ is a binary classfication task. Cross-entropy loss is used for each task. Therefore, the total loss in our proposed boundary-aware FCN is
where \(\theta =\left\{ \theta _{r}, \theta _{b}, \theta _{f} \right\} \) is the set of weight parameters in the boundary-aware FCN. \(\mathcal {L}_{t}\) refers to the loss function of each task. \(x_{n,i}\) is the i-th voxel in the n-th image used for training, and \(P_{t}\) refers to the predicted probability of the voxel \(x_{n,i}\) belonging to class \(l_{t}\). Similarly to [15], we extract boundaries from radiologists’ region annotations and dilate them with a disk filter.
In the boundary-aware FCN, 2D axial slices from 3D MR volumes are used as input. In addition, since adding brain symmetry information is helpful for FCN based tumor segmentation [8], symmetric intensity difference maps are combined with original slices as input, resulting in 8 input channels (see Figs. 2 and 3).
4 Evaluation
Our model was evaluated on BRATS13 and BRATS15 datasets. BRATS13 contains 20 HG patients for training and 10 HGs for testing. (The 10 low-grade patients were not used.) From BRATS15, we used 220 annotated HG patients’ images in the training set. For each patient there were 4 modalities (T1, T1-contrast (T1c), T2 and Flair) which were skull-stripped and co-registered. Quantitative evaluation was performed on three sub-tasks: (1) the complete tumor (including all four tumor structures); (2) the tumour core (including all tumor structures except edema); (3) the enhancing tumor region (including only the enhancing tumor structure).
Our model was implemented with the Keras and Theano backend. For each MR image, voxel intensities were normalised to have zero mean and unit variance. Networks were trained with back-propagation using Adam optimizer. Learning rate was 0.001. The downsampling path was initialized with VGG-16 weights [7]. Upsampling paths were initialized randomly using the strategy in [17].
4.1 Results on BRATS15 Dataset
We randomly split HG images in the BRATS15 training set into three subsets at a ratio of 6:2:2, resulting in 132 training, 44 validation and 44 test images. Three models were compared: (1) variant of FCN (Fig. 2), denoted FCN; (2) FCN with a fully-connected CRF [6]; (3) the multi-task boundary-aware FCN.
Firstly, FCN models were evaluated on the validation set during training. Figure 4(a) plots Dice values for the Complete tumor task for boundary-aware FCN and FCN. Using boundary-aware FCN improved performance at most training epochs, giving an average 1.1% improvement in Dice. No obvious improvement was observed for Core and Enhancing tasks. We further performed a comparison by replacing the combination stage with the threshold-based fusion method in [15]. This resulted in Dice dropping by 15% for the Complete tumor task (from 88 to 75), which indicates the combination stage was beneficial. We experimented adding more layers to FCN (e.g., using four convolutional blocks in downsampling path and four upsampling paths) but observed no improvement, suggesting the benefit of boundary-aware FCN is not from simply having more layers or parameters.
The validation performance of both models saturated at around 30 epochs. Therefore, models trained at 30 epochs were used for benchmarking on test data. On the 44 unseen test images, results of boundary-aware FCN, single-task FCN and FCN+CRF are shown in Table 1. The boundary-aware FCN outperformed FCN and FCN+CRF in terms of Dice and Sensitivity but not in terms of Positive Predictive Value.
One advantage of our model is its improvement of tumor boundaries. To show this, we adopt the trimap [6] to measure precision of segmentation boundaries for complete tumors. Specifically, we count the proportion of pixels misclassified within a narrow band surrounding tumor boundaries obtained from the experts’ ground truth. As shown in Fig. 4(b), boundary-aware FCN outperformed single-task FCN and FCN+CRF across all trimap widths. For each trimap width used, we conducted a paired t-test over the 44 pairs, where each pair is the performance values obtained on one validation image by boundary-aware FCN and FCN. Small p-values (p < 0.01) in all 7 cases indicate that the improvements are statistically significant irrespective of the trimap measure used. Example segmentation results for boundary-aware FCN and FCN are shown in Fig. 5. It can be seen that boundary-aware FCN removes both false positives and false negatives for the complete tumor task.
We conducted another experiment without using symmetry maps. Boundary-aware FCN gave an average of 1.3% improvement in Dice compared to FCN. The improvement for boundaries was statistically significant (p < 0.01).
4.2 Results on BRATS13 Dataset
A 5-fold cross validation was performed on the 20 HG images in BRATS13. Training folds were augmented by scaling, rotating and flipping each image. Performance curves for Dice and trimap show similar trends as for BRATS15 (Fig. 4(c)–(d)). However, using CRF did not improve performance on this dataset, suggesting boundary-aware FCN is more robust in improving boundary precision. The improvement of trimap is larger than for BRATS15. It is worth noting that, in contrast to BRATS15 (where ground truth was produced by algorithms, though verified by radiologists), the ground truth of BRATS13 is the fusion of annotations from multiple radiologists. Thus the improvement gained by our method on this set is arguably more solid evidence showing the benefit of joint learning, especially on improving boundary precision.
Our method is among the top-ranking on the BRATS13 test set (Table 2). Tustison et al. [2], the winner of BRATS13 challenge [1], used an auxiliary health brain dataset for registration to calculate the asymmetry features, while we only use the data provided by the challenge. Our model is fully automatic and overall ranked higher than a semi-automatic method [9].
Regarding CNN methods, our results are competitive with Pereira et al. [3] and better than Havaei et al. [4]. Zhao et al. [10] applied joint CNN with CRF training [18]. Our boundary-aware FCN gave better results without the cost of tuning a CRF. A direct comparison with 3D CNN is not reported here as Kamnitsas et al. [5] did not report results on this dataset.
One advantage of our model is its relatively low computational cost for a new test image. Kwon et al. [9] reported an average running time of 85 min for each 3D volume on a CPU. For CNN approaches, Pereira et al. [3] reported an average running time of 8 min while 3 min was reported by Havaei et al. [4], both using a modern GPU. For an indicative comparison, our method took similar computational time to Havaei et al. [4]. Note that, in our current implementation, 95% of the time was used to compute the symmetry inputs on CPU. Computation of symmetry maps parallelized on GPU would provide a considerable speed-up.
5 Conclusion
We introduced a boundary-aware FCN for brain tumor segmentation that jointly learns boundary and region tasks. It achieved state-of-the-art results and improved the precision of segmented boundaries on both BRATS13 and BRATS15 datasets compared to the single-task FCN and FCN+CRF. It is among the top ranked methods and has relatively low computational cost at test time.
References
Menze, B.H., Jakab, A., Bauer, S., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). Med. Imaging 34(10), 1993–2024 (2015)
Tustison, N.J., Shrinidhi, K.L., Wintermark, M., et al.: Optimal symmetric multimodal templates and concatenated random forests for supervised brain tumor segmentation (simplified) with ANTsR. Neuroinformatics 13(2), 209–225 (2015)
Pereira, S., Pinto, A., Alves, V., et al.: Brain tumor segmentation using convolutional neural networks in MRI images. Med. Imaging 35(5), 1240–1251 (2016)
Havaei, M., Davy, A., Warde-Farley, D., et al.: Brain tumor segmentation with deep neural networks. Med. Image Anal. 35, 18–31 (2017)
Kamnitsas, K., Ledig, C., Newcombe, V.F., et al.: Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017)
Krähenbühl, P., Koltun, V.: Efficient inference in fully connected CRFs with Gaussian edge potentials. In: NIPS, pp. 109–117 (2011)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
Shen, H., Zhang, J., Zheng, W.: Efficient symmetry-driven fully convolutional network for multimodal brain tumor segmentation. In: ICIP (2017, to appear)
Kwon, D., Shinohara, R.T., Akbari, H., Davatzikos, C.: Combining generative models for multifocal glioma segmentation and registration. In: Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R. (eds.) MICCAI 2014. LNCS, vol. 8673, pp. 763–770. Springer, Cham (2014). doi:10.1007/978-3-319-10404-1_95
Zhao, X., Wu, Y., Song, G., et al.: Brain tumor segmentation using a fully convolutional neural network with conditional random fields. In: Crimi, A., Menze, B., Maier, O., Reyes, M., Winzeck, S., Handels, H. (eds.) BrainLes 2016. LNCS, pp. 75–87. Springer, Cham (2016). doi:10.1007/978-3-319-55524-9_8
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: CVPR, pp. 3431–3440 (2015)
Chen, L.C., Papandreou, G., Kokkinos, I., et al.: Semantic image segmentation with deep convolutional nets and fully connected CRFs. arXiv preprint arXiv:1412.7062 (2014)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). doi:10.1007/978-3-319-24574-4_28
Chen, H., Qi, X.J., Cheng, J.Z., Heng, P.A.: Deep contextual networks for neuronal structure segmentation. In: AAAI (2016)
Chen, H., Qi, X., Yu, L., Heng, P.A.: DCAN: deep contour-aware networks for accurate gland segmentation. In: CVPR, pp. 2487–2496 (2016)
Xu, Y., Li, Y., Liu, M., Wang, Y., Lai, M., Chang, E.I.-C.: Gland instance segmentation by deep multichannel side supervision. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 496–504. Springer, Cham (2016). doi:10.1007/978-3-319-46723-8_57
He, K., Zhang, X., Ren, S., Sun, J.: Delving deep into rectifiers: surpassing human-level performance on imagenet classification. In: ICCV, pp. 1026–1034 (2015)
Zheng, S., Jayasumana, S., Romera-Paredes, B., et al.: Conditional random fields as recurrent neural networks. In: ICCV, pp. 1529–1537 (2015)
Acknowledgments
This work was supported partially by the National Natural Science Foundation of China (No. 61628212).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Shen, H., Wang, R., Zhang, J., McKenna, S.J. (2017). Boundary-Aware Fully Convolutional Network for Brain Tumor Segmentation. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds) Medical Image Computing and Computer-Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science(), vol 10434. Springer, Cham. https://doi.org/10.1007/978-3-319-66185-8_49
Download citation
DOI: https://doi.org/10.1007/978-3-319-66185-8_49
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-66184-1
Online ISBN: 978-3-319-66185-8
eBook Packages: Computer ScienceComputer Science (R0)