Abstract
Cardiac magnetic resonance (CMR) can be used for quantitative analysis of heart function. However, CMR imaging typically involves acquiring 2D image planes during separate breath-holds, often resulting in misalignment of the heart between image planes in 3D. Accurate quantitative analysis requires a robust 3D reconstruction of the heart from CMR images, which is adversely affected by such motion artifacts. Therefore, we propose a fully automated method for motion correction of CMR planes using segmentations produced by fully convolutional neural networks (FCNs). FCNs are trained on 100 UK Biobank subjects to produce short-axis and long-axis segmentations, which are subsequently used in an iterative registration algorithm for correcting breath-hold induced motion artifacts. We demonstrate significant improvements in motion-correction over image-based registration, with strong correspondence to results obtained using manual segmentations. We also deploy our automatic method on 9,353 subjects in the UK Biobank database, demonstrating significant improvements in 3D plane alignment.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Cardiac magnetic resonance (CMR) is an established clinical imaging technique for the assessment of cardiovascular disease. Three-dimensional (3D) reconstruction from multiplanar short-axis (SA) and, orthogonal to these, long-axis (LA) cine CMR can be used for the quantitative analysis of heart function, including volumetric measurements [7] and cardiac motion [1].
Current clinical protocols typically require a subject to take a separate breath-hold for the acquisition of each image plane over the cardiac cycle. Variation in breath-hold positions for the acquisition of different planes alters the position of the heart with respect to the scanner, resulting in misalignment of the cardiac geometry between image planes. These motion artifacts can cause errors in the 3D reconstruction of relevant cardiac structures from these image planes including the left-ventricle (LV) myocardium and blood-pool, and can therefore adversely affect the subsequent analysis of heart function.
CMR Motion Artifacts: CMR image acquisition is typically gated by an electrocardiogram (ECG). ECG-gating allows for temporal frames to be acquired at corresponding phases of the cardiac cycle for each image plane. An implication of ECG gating is that the contractile state and resulting shape of the heart is fixed for each respective phase of the cardiac cycle. As a result, the heart undergoes an approximately rigid transformation between different breath-hold positions, and mainly a translation in the craniocaudal direction [5]. However, the field of view of standard CMR images typically includes surrounding features within and including the rib cage. Taken as a whole, these features undergo a non-rigid deformation between different breath-holds. Therefore, when applying rigid registration to align standard LA and SA images, the non-rigid deformation of the image content under different breath-holds is not adequately modelled and can result in poor alignment of the myocardium.
Related Work: Previous work to correct breath-hold induced motion artifacts includes the minimisation of a global cost function by rigidly registering together 2D LA and SA images [4], or their local phase transform [12]. However, without cropping the images to a tighter region around the heart, this approach suffers from the problem of non-rigid deformation of features surrounding the heart. Another approach has been to register 2D images to a simultaneously acquired 3D image volume to take into account through-plane rotations and translations [13]. This approach is also susceptible to non-rigid deformation of features around the heart under different breath-holds, and requires a time-consuming 3D scan. Mesh-based methods have enforced smoothness of a 3D mesh fitted to SA myocardial contours [10], although this approach disregards the 3D anatomical information provided by combining LA and SA images. For 3D/3D registration of cardiac CT volumes, identification of cardiac geometry has been used to overcome limitations of image-to-image registration [6].
Contributions: Intuitively, if the geometry of the heart at a given cardiac phase can be accurately identified in the LA and SA views, then a rigid registration could be applied to optimize alignment of the 3D heart geometry from all images, without issues arising due to non-rigid deformation of background features. A common representation of cardiac anatomy is the use of segmentation labels. Typically, manual segmentation is required to produce labels, but processing large datasets in this way can be prohibitively time-consuming and prone to inter-observer error. To overcome this problem, Fully Convolutional Neural Networks (FCNs) have recently produced state-of-the-art results in cardiac segmentation [11]. In this paper we present a method for breath-hold induced motion artifact correction using automatic myocardial segmentations as the input to a rigid registration algorithm. Specifically we present:
-
FCNs trained separately on LA and SA images to produce high accuracy labels of LV myocardium and blood-pool in multiple image planes;
-
An iterative algorithm to correct motion artifacts by aligning LA and SA segmentations of the LV myocardium and blood-pool;
-
A comparison of automatic segmentation-based alignment, manual segmentation-based alignment, and image-based alignment on UK Biobank data.
2 Materials and Methods
2.1 Image Data
SA and LA CMR images were used from a subset of the UK Biobank databaseFootnote 1 (see [7] for full CMR protocol). Typically 10–12 SA planes and, orthogonal to these, three LA planes comprising the 2-chamber (2Ch), 3-chamber (3Ch) and 4-chamber (4Ch) views were available for each subject. The SA images have an in-plane resolution of 1.8 mm and slice thickness of 8 mm, while the LA images have in-plane resolution of 1.8 mm and slice thickness of 6 mm. The SA and LA views were segmented (see Sect. 3.1) producing a LV blood-pool label = 1, myocardium label = 2, and background label = 0. Note that the right ventricle was excluded since its inclusion generally produced worse results for the registration due to greater segmentation ambiguities caused by partial-volume effects.
2.2 Segmentation Network
Deep convolutional neural networks (CNNs) have emerged in recent years as a powerful method to learn image features for tasks such as image classification [9] and segmentation [3, 8]. Networks used for image classification such as the VGG-net [9] learn image features through cascaded layers of increasingly coarse feature maps connected via combinations of convolutions and pooling operators, with an output dimensionality equal to the number of image classes. The FCN [3] and U-net [8] architectures, on the other hand, produce pixel-wise predictions of image labels by up-sampling coarse feature maps from different levels of the network to an output with the resolution of the input images.
Problem Formulation: Let x be an image and y be its corresponding pixel-wise label map, where a training set S consists of pairs of images and label maps, \(S = \{x_i | i = 1,2,\dots ,N ; y_i | i = 1,2,\dots ,N\}\). Supervised learning is performed to estimate the network parameters, \(\varTheta \), to predict label map \(y_i\) of image \(x_i\) in the training set, by optimising the cross-entropy loss function
where j denotes the pixel index and \(P(y_{i,j} | x_i, \varTheta )\) denotes the softmax probability produced at pixel j for image (and corresponding label map) i.
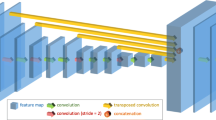
Network Architecture: A VGG-like network with FCN architecture is used for the automatic segmentation of the LV myocardium and blood-pool, as shown in Fig. 1. Batch-normalisation (BN) is used after each convolutional layer, and before a rectified linear unit (ReLU) activation. The BN operation removes internal covariate shift, improving training speed.
As shown in Fig. 1, input images have pixel dimensions \(224\times 224\). Every layer in the diagram prefixed by “C" performs the operation: convolution \(\rightarrow \) BN \(\rightarrow \) ReLU, with the exception of C15 and C17. The (filter size/stride) is \((3\times 3/1)\) for layers C1 to C13, except for layers C3, C5, C8 and C11 which have \((3\times 3/2)\). The arrows represent \((3\times 3/1)\) convolutional layers (\(C14a-e\)) followed by a bilinear up-sampling (up) layer with a factor necessary to achieve feature map volumes with size \(224\times 224\times 64\), all of which are concatenated into the orange feature map volume. C15 and C16 are \((1\times 1/1)\) convolutional layers, where C15 performs convolution \(\rightarrow \) linear activation. C17 applies a \((1\times 1/1)\) convolution with a pixel-wise softmax activation, producing the green feature map volume with a depth = 3, corresponding to the number of image labels.
2.3 Multiplanar Registration Algorithm
An iterative algorithm was developed for the registration of SA and LA images in 3D using the automatic segmentations. Similarly to [12], a global convergence is reached by iteratively registering each image plane to its intersection with the other image planes, which are kept fixed in space. Each cycle over all N image planes (from both SA and LA views) constitutes one iteration.
Registering segmentations is advantageous since it allows a the use of a computationally efficient similarity measure, such as the Sum of Squared Differences (SSD). Let \(S_{i,j}\) denote the segmented image at slice i and iteration j, and let \(\hat{S}_{i,j}\) denote the segmented image computed from the other image planes intersecting with \(S_{i,j}\). Nearest-neighbour interpolation of the intersecting planes was used in order to preserve the segmentation labels of the myocardium and blood-pool. Gradient descent was used to minimise SSD,
where k indexes the image pixels, and \(\varPhi _{i,j} = \{\varDelta x_{i,j}, \varDelta y_{i,j}\}\) are the translation parameters applied to transform \(S_{i,j}\). Only in-plane translations were considered as minimal in-plane rotation was assumed [5]. Additionally, through-plane translation/rotation was assumed to be minimal relative to the slice thicknesses, which were 8 mm and 6 mm for the SA and LA planes, respectively [5].
To preserve the segmentation labels at each iteration, the translations produced in Eq. 2 are rounded to the nearest whole-pixel increment. We denote these whole-pixel translations by \(\varPhi ^p_{i,j} = \{\varDelta x^p_{i,j}, \varDelta y^p_{i,j}\}\). Under this constraint, the above procedure was performed until M iterations where the translation norm of all planes, N, summed to zero,
At this point, the final translation for slice i from its original position was computed as the summation over all iterations,
which was used to align the original CMR images and segmentations. Iterating over slice-wise sub-problems in this way to find the final translation parameters for each slice, i, is akin to finding a global minimum [2] for a cost function:
3 Experiments and Results
3.1 Segmentation
Pre-processing: A set of 100 subjects was randomly sampled from the UK Biobank database and expert manual segmentations of the LA and SA planes were created in ITK-SNAPFootnote 2 with labels described in Sect. 2.1. For the SA planes, approximately 1000 end-diastolic (ED) SA images were segmented, as well as 100 ED LA images in each LA view. All images and labels were reshaped to \(224\times 224\) pixels by zero-padding. Image pixel dimensions varied between subjects, with a mean of 190 pixels, a maximum of 210 and a minimum of 132.
Training: Separate networks (using the architecture in Fig. 1) were trained for the (i) SA, (ii) 2Ch, (iii) 3Ch, and (iv) 4Ch views. The data were split into \(80\%\) training and \(20\%\) validation datasets. Mini-batches were used to train each network with batch sizes of 20 and 5 for (i) and (ii)–(iv), respectively. On-the-fly data augmentation was used for each mini-batch, with random rotation, scaling and translation applied to the images. Training was run with a learning rate of 0.001 for 500 epochs after which the change in validation accuracy was negligible.
Assessment: Dice score was used to compare the overlap of the generated labels to the ground truth labels in the validation data. Furthermore, intra-observer variability was assessed by comparing the manual validation segmentations to a second round of segmentations performed by the same expert.
Results: In Table 1, the intra-observer Dice scores are slightly higher than that of the FCN output for both labels, by approximately \(1\%\) in both SA and LA views. This demonstrates the FCNs’ consistency in segmenting both the myocardium and blood-pool in all views. Performance of FCNs trained on the LA views was similar to that trained on the SA view despite far fewer images in the training set. This is likely due to the greater variation in SA image content, which varies from base to apex planes. The blood-pool Dice score was higher for the LA views compared to SA. Absence of papillary muscles in the 4Ch view may explain the higher myocardium Dice score than the 2Ch and 3Ch views.
3.2 Motion Correction
Pre-processing: Two additional sets of 20 subjects were curated from the UK Biobank database to assess the motion correction algorithm. The first 20 subjects (Set 1) were selected based on visual inspection for having LA and SA images with negligible motion artifacts. The second 20 subjects (Set 2) were selected based on having visibly moderate to severe motion artifacts.
Performance Assessment: A comparison was made between four methods: (a) no registration; (b) image-based (IB) registration; (c) segmentation-based registration using manual segmentation (SB-manual); and (d) segmentation-based registration using the FCN output (SB-FCN). Results for each method on Set 1 and Set 2 are shown in Table 2. For the IB registration, the same iterative approach was used as described in Sect. 2.3, except with a Normalised Mutual Information similarity measure on image intensities, and a maximum number of iterations set to \(M=5\). Convergence of SB registrations was typically achieved within 3 iterations. The mean distance in pixels between the endocardial contours (MCD) from the LA versus SA planes was used to assess the registration results of the different methods. Note that manual segmentations were compared for (a)–(c), and the FCN segmentations were used for (d). Additionally, the SB-FCN method was assessed on 9,353 subjects with CINE MRI scans in the UK Biobank database (‘UKBB’ in Table 2).
To assess the smoothness and anatomical accuracy of the 3D reconstruction, a statistical shape model (SSM) LV mesh was fitted to the SA myocardium segmentations in 3D before and after alignment [1]. The Dice score between the fitted SSM and SA LV myocardium segmentation is a measure of integrity for further quantitative analysis, for example to analyse 3D motion.
For comparisons within Set 1, Set 2 and UKBB, a two-tailed paired t-test was used to determine significant differences between normally distributed samples, and the Wilcoxon signed ranked test for non-normally distributed samples. For Comparison between Set 1 and Set 2, a two-tailed unpaired t-test and the Mann-Whitney U test were used for normally and non-normally distributed samples, respectively. Normality was tested using the Shapiro-Wilk test.
Results: Significant differences are reported for \(p<0.01\). Referring to Table 2, for Dice scores in Set 1, there were significant differences between both (a) and (c) compared to (d) (\(^*\)). There were no significant differences in MCD between them, however, suggesting similar alignment of Set 1 using all methods. The lower Dice score of the SB-FCN method is addressed in the Sect. 4.
There was both a significant increase in MCD and decrease in Dice score for methods (a) and (b) on Set 2 (†) compared to (a) and (b) on Set 1 as well as (c) and (d) on Set 2. Conversely, there was no significant difference in MCD and Dice score between (c) and (d) on Set 2 (bold) compared to (c) and (d) on Set 1. This suggests that (c) and (d) produce as good results on Set 2 as they do on Set 1, demonstrating their ability to correct moderate to severe artifacts. Figure 2 shows results for a case with severe motion artifacts, where method (b) fails to align the 3D geometry, but both (c) and (d) succeed. Finally for UKBB, SB-FCN (\(^{**}\)) significantly improved MCD and Dice scores compared to no alignment.
4 Discussion and Conclusions
We have proposed a fully automated method for the robust correction of breath-hold induced motion artifacts using FCNs and an iterative registration algorithm. The FCN segmentations approached expert intra-observer Dice scores for both SA and LA views. The proposed SB-FCN registration produces significantly better results than IB registration for cases with moderate to severe motion artifacts, and is considerably faster than a SB-manual approach. The SB-FCN method also significantly improves 3D alignment of CMR planes and segmentations for a large dataset of 9,353 subjects.
A limitation of our method is that the FCN struggles to accurately segment the most apical and basal SA slices with myocardium, leading to slightly lower Dice scores compared to the SB-manual method in Table 2. In future work, networks could be trained specifically for the SA apex and base images, and segmentations and images could be used jointly for registration. Our approach can also be easily adapted for data from different scanners via transfer learning.
References
Bai, W., Shi, W., De Marvao, A., et al.: A cardiac atlas built from high resolution MR images of 1000 + normal subjects and atlas-based analysis of cardiac shape and motion. Med. Image Anal. 26(1), 133–145 (2015)
Bezdek, J., Hathaway, R.: Convergence of alternating optimization. Neural, Parallel Sci. Comput. 11(4), 351–368 (2003)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: CVPR, pp. 3431–3440 (2015)
Lötjönen, J., Pollari, M., Kivistö, S., Lauerma, K.: Correction of movement artifacts from 4-D cardiac short- and long-axis MR data. In: Barillot, C., Haynor, D.R., Hellier, P. (eds.) MICCAI 2004. LNCS, vol. 3217, pp. 405–412. Springer, Heidelberg (2004). doi:10.1007/978-3-540-30136-3_50
McLeish, K., Hill, D., Atkinson, D., et al.: A study of the motion and deformation of the heart due to respiration. IEEE Trans. Med. Imaging 21(9), 1142–1150 (2002)
Neumann, D., Grbi, S., John, M., et al.: Probabilistic sparse matching for robust 3D/3D fusion in minimally invasive surgery. IEEE Trans. Med. Imaging 34(1), 49–60 (2015)
Petersen, S., Matthews, P., Francis, J., et al.: UK Biobank’s cardiovascular magnetic resonance protocol. J. Cardiovasc. Magn. Reson. 18(1), 8 (2016)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). doi:10.1007/978-3-319-24574-4_28
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. In: ICLR (2014)
Su, Y., Tan, M.L., Lim, C.W., et al.: Automatic correction of motion artifacts in 4D left ventricle model reconstructed from MRI. Comput. Cardiol. 41, 705–708 (2014)
Tran, P.: A Fully Convolutional Neural Network for Cardiac Segmentation in Short-Axis MRI. CoRR (2016). http://arxiv.org/abs/1604.00494
Villard, B., Zacur, E., Dall’Armellina, E., Grau, V.: Correction of slice misalignment in multi-breath-hold cardiac MRI scans. In: Mansi, T., McLeod, K., Pop, M., Rhode, K., Sermesant, M., Young, A. (eds.) STACOM 2016. LNCS, vol. 10124, pp. 30–38. Springer, Cham (2017). doi:10.1007/978-3-319-52718-5_4
Zakkaroff, C., Radjenovic, A., Greenwood, J., Magee, D.: Stack alignment transform for misalignment correction in cardiac MR cine series. Technical report, University of Leeds (2012)
Acknowledgements
This work was funded by EPSRC grants EP/K030310/1 and EP/K030523/1. This research was conducted using the UK Biobank resource under Application Number 17806. The Titan X used for this research was donated by the NVIDIA Corporation.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Sinclair, M., Bai, W., Puyol-Antón, E., Oktay, O., Rueckert, D., King, A.P. (2017). Fully Automated Segmentation-Based Respiratory Motion Correction of Multiplanar Cardiac Magnetic Resonance Images for Large-Scale Datasets. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds) Medical Image Computing and Computer-Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science(), vol 10434. Springer, Cham. https://doi.org/10.1007/978-3-319-66185-8_38
Download citation
DOI: https://doi.org/10.1007/978-3-319-66185-8_38
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-66184-1
Online ISBN: 978-3-319-66185-8
eBook Packages: Computer ScienceComputer Science (R0)