Abstract
Advances in genomics have driven to the recognition that tumours are populated by different minor subclones of malignant cells that control the way the tumour progresses. However, the spatial and temporal genomic heterogeneity of tumours has been a hurdle in clinical oncology. This is mainly because the standard methodology for genomic analysis is the biopsy, that besides being an invasive technique, it does not capture the entire tumour spatial state in a single exam. Radiographic medical imaging opens new opportunities for genomic analysis by providing full state visualisation of a tumour at a macroscopic level, in a non-invasive way. Having in mind that mutational testing of EGFR and KRAS is a routine in lung cancer treatment, it was studied whether clinical and imaging data are valuable for predicting EGFR and KRAS mutations in a cohort of NSCLC patients. A reliable predictive model was found for EGFR (AUC = 0.96) using both a Multi-layer Perceptron model and a Random Forest model but not for KRAS (AUC = 0.56). A feature importance analysis using Random Forest reported that the presence of emphysema and lung parenchymal features have the highest correlation with EGFR mutation status. This study opens new opportunities for radiogenomics on predicting molecular properties in a more readily available and non-invasive way.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Lung cancer is the most common cause of cancer death in the world, responsible for nearly 1.6 million deaths annually [10]. The main contributing factor for the high death rate of lung cancer is the late diagnosis [19]. Once diagnosed, lung cancer is often in an advanced stage, with 15% or less chance of a 5-year survival [15]. At that stage, tumours are already composed by multiple clonal subpopulations of cancer cells and, consequently, the treatment must be shaped based on the individual tumour heterogeneity. Precision medicine is the medical field that tailors practices and/or therapies to individual patients by taking into account the individual variability of genes. The traditional method of analysing the tumour is by extracting tumour tissue in a biopsy, which is then characterised using genomic-based approaches. In spite of being a successful approach in clinical oncology, repeated biopsies tend to increase medical complications. Further, tumour characterisation usually demands several biopsies, since the results can vary depending on the part of the tumour that is analysed [23].
In Non-small cell lung cancer (NSCLC), which accounts 85% of all lung cancers [5], mutational testing of selected genes is a standard practice to determine how affected patients will respond to targeted therapy [9]. This includes determining the mutation status of epidermal growth factor receptor (EGFR), a cell receptor that activates growth and survival [24], and Kristen rat sarcoma viral oncogene homolog (KRAS), which activates the same pathway as EGFR when mutated [21]. Patients with mutant EGFR are sensitive to tyrosine kinase inhibitors (TKIs) gefitinib and erlotinib. Hence, patients with mutated EGFR lung cancer, who receive treatments with targeted TKIs are expected to have a longer progression-free survival in comparison to chemotherapy treatment. However, if gefitinib is administered in cases with non-mutated EGFR, the patient will undergo a shorter progression-free survival [18]. KRAS mutation status is also helpful for treatment planning. It has proven to be correlated with response to chemotherapy since patients with mutated KRAS which undergo chemotherapy have revealed inferior responses and shorter survival compared to patients with no KRAS mutation [13, 26]. On that basis, identifying patients with mutated EGFR and KRAS is highly important in precision medicine.
As a less invasive technique compared to biopsy, radiographic medical imaging opens new opportunities for tumour characterisation. Images exhibit strong phenotypic differences between tumours, such as tumour size, presence of emphysema and/or fibrosis. Those differences normally fail to be recognised by the naked eye, thus they may have the potential to be valuable predictors of therapeutic benefit. Moreover, a great advantage of medical imaging is its ability to provide a full state visualisation of a tumour at a macroscopic level. Therefore, radiogenomics, the fusion of medical images and genomics, offers attractive opportunities for non-invasive treatment planning.
Given the relevance of the problem, in this paper, we propose predictive models for EGFR and KRAS mutation status, using a set of clinical and radiologist-observed qualitative imaging features, taking advantage of the learning capabilities of machine learning techniques.
The remainder of this paper is as follows. In Sect. 2, we present the related work which has been done so far. In Sect. 3, we present the proposed approach, while in Sect. 4 we detail and discuss the experimental results. We conclude the paper with Sect. 5, summarising its main contributions and findings.
2 Related Work
A thorough search of the relevant literature yielded only one related article which investigated whether EGFR and KRAS mutation status can be predicted using qualitative features obtained from imaging data. Gevaert et al. [11] used 89 qualitative image features of NSCLC patients tumours, annotated by a thoracic oncologist, to create models to predict EGFR and KRAS mutation status. A univariate correlation study was performed between mutation status and the qualitative imaging features and, afterwards, the most correlated features were used in a multivariate analysis using decision trees. Emphysema, airway abnormality, the percentage of ground glass component and the type of tumour margin reached the significance threshold of correlation with EGFR mutation status and they were used to build a decision tree model, which achieved an area under the ROC curve (AUC) of 0.89. With regard to KRAS mutation, no features reached the significance threshold of correlation and, consequently, the models built for KRAS were not considered useful (AUC = 0.55). Furthermore, some studies have investigated the association between EGFR mutation status and quantitative features, rather than qualitative features [7, 16, 17].
With a view on the advances that have been made, in this study, we propose different experimental methodologies that take advantage of powerful machine learning techniques to create predictive models using a new set of features. By using a different cohort of patients, as well as different features, one can further evaluate the relation between EGFR and KRAS mutation status and radiographic imaging data.
3 Methodology
This study aimed to investigate whether clinical and qualitative imaging features are advantageous mutation status predictors and build predictive models using two algorithms: Random Forest (RF) [3] and Multi-layer Perceptrons (MLP) [22] networks. The data was divided into a training set (80%) and a test set (20%), ensuring that each set maintains an equal proportion of instances of each class. It is important to mention that the set of subjects used for training and testing were kept constant for all the performed experiments. Hyper-parameters were chosen applying grid-search with 5-fold cross-validation to the training data and selecting the set of hyper-parameters of the model with the highest F-measure. The developed code and used data are available on Github.Footnote 1
3.1 The Dataset
The study included a subset of 158 NSCLC patients tested for EGFR mutation status and 157 NSCLC patients tested for KRAS mutation status, characterised by qualitative and clinical features. The data was obtained from the open-access NSCLC-Radiogenomics dataset available at the cancer imaging archive (TCIA) database [2, 6, 12].
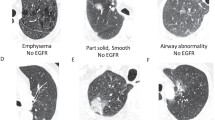
The qualitative features were obtained from an analysis of pre-treatment computed tomography (CT) images using a controlled vocabulary. The used terms are commonly used in radiology clinical practice and derive from descriptions in the radiology literature [1]. Definitions of some of the terms used in this description can be found in [14]. The template of semantic terms was developed exclusively for nodules since it is the most prevalent expression of lung cancer. Therefore, other manifestations of lung cancer besides nodules (e.g. central obstructive tumours) are not included in this study.
From the 30 qualitative features available in the NSCLC-Radiogenomics dataset, some were discarded due to a large number of not applicable values (e.g. the fibrosis type field in a patient that has fibrosis absent), thus, a subset of 18 qualitative features was used in this study. The used set includes nodule and parenchymal features, which describe the nodules geometry, location, internal features and other related findings. Additionally, the patient’s gender and smoking status were considered due to its significant association with mutation status prevalence, confirmed in recent studies [8, 20, 25]. From this point forward, gender and smoking status are designated as clinical features and the qualitative features extracted from the images as imaging features. Table 1 shows detailed information regarding the data distribution and the nomenclature used to classify the tumours.
The dataset comprises percentages of 26% and 25% mutated cases for EGFR and KRAS, respectively. Before feeding the data into the model, features were converted to binary vectors following a one-hot encoding strategy. Thereafter, the number of features increased from 20 to 73.
3.2 Random Forest
RF models were implemented for predicting EGFR and KRAS mutation status. As an algorithm based on ensemble learning, RF makes the predictions taking advantage of a group of models, instead of a single model. Random Forest samples both observations and features of training data in order to build independent decision trees which contribute by voting for the ensemble prediction. Bearing in mind that a decision tree is an unstable algorithm, by averaging the results of all decision trees, the variance component of the model will be minimised, which approximates the ensemble to an ideal model.
Due to the ability of RF to recognise the importance of the features for the problem in mind, it was conducted an analysis of the most valuable ones.
3.3 Multi-layer Perceptron
By virtue of the remarkable ability of MLP to extract patterns and detect trends, its performance on predicting the genes mutation status was tested.
The MLP assumes the distribution of classes is similar, which in this case would result in a model biased towards the negative class. However, in this study, the correct classification of both classes is equally important, since the classification of a patient with the wrong mutation status could lead to the administration of a less suitable treatment and, consequently, to shorter progression-free survival. To overcome class imbalance, it was conducted a Synthetic Minority Over-sampling Technique for Nominal and Continuous (SMOTE-NC) data approach, in which new instances are created based on the 5 nearest neighbours of the feature space that belong to the minority class. In comparison to traditional over-sampling, SMOTE-NC has the advantage of building a more general decision region of the minority class [4]. After applying SMOTE-NC, the training set contains the same number of mutated and wildtype samples. On the premise of keeping the fed data constant among the two algorithms, SMOTE-NC was applied to all the performed experiments.
4 Results and Discussion
The same experimental set-up was followed regarding EGFR and KRAS mutations. Interestingly, the average results were quite different, in a sense that it was possible to achieve models that reliably predict EGFR mutations but not KRAS mutation presence. From the experiments conducted, the predictive model for KRAS mutation with the best performance had an AUC of 0.56, having the RF as the classifier. Therefore, the following results are exclusively regarding EGFR.
4.1 EGFR Mutation Status Prediction
Three experiments were conducted in order to achieve the greatest set of features to predict EGFR mutation status: using clinical features, imaging features and both imaging and clinical features. Clinical features were only attempted with MLP, since the RF is not an ideal model for a set of 2 features, due to the lack of feature combinations. The range of values used in the grid search for RF and MLP for each hyper-parameter is presented in Tables 2 and 3, respectively.
The hyper-parameters of the models which achieved the highest F-measure in the 5-fold cross-validation for RF and MLP are included in Tables 4 and 5, respectively, for each experiment.
Table 6 shows the results obtained on the test set by the RF and MLP models which provided the best results in the training set, in the three performed experiments.
On average, a better performance was achieved when both clinical and imaging features were used. The RF model achieved an equal performance using both clinical and imaging features or just imaging features; however, MLP reached a higher performance (AUC = 0.96) when clinical features were added to the feature set.
Focusing on the MLP experiments, clinical features achieved a satisfactory performance when predicting EGFR mutation status (AUC = 0.68), whereas, when using imaging data a reliable predictive model is obtained (AUC = 0.94). Further, when imaging and clinical data were combined, it was created a model which further increases the imaging data performance (AUC = 0.96). The RF model achieved the same performance in the two experiments potentially due to its intrinsic feature subsampling.
The ROC curves for the RF and MLP models using imaging and clinical features are presented in Fig. 1. The confusion matrix obtained by the two models using imaging and clinical features was identical, and it is presented in Fig. 2.
Feature Importance. Taking advantage of the RF ability to recognise features importance, it was conducted an analysis in order to find the most valuable predictors amongst the used set of features. The RF model outputs a score for each feature which sums to one, and it describes the average decrease in impurity over trees.
Since the one-hot encoding approach was used to convert from categorical data to binary vectors, there is an importance score associated with each feature value and not a single score to the feature itself. Therefore, the feature score was considered to be the sum of the scores of each of its values. For instance, if having emphysema present had a score of 0.10 and emphysema absent a score of 0.08, the feature emphysema has a score of 0.18. Figure 3 shows the importance scores as a result of this analysis. Emphysema is plainly the feature with the highest score (0.18) followed by lung parenchymal features (0.16).
5 Conclusions and Future Work
In this work, we report a radiogenomics model that is able to predict the mutation status of EGFR (AUC = 0.96) from CT scans in a less invasive procedure, compared to repeated biopsies during treatment. To the best of our knowledge, this work was the second attempt to create predictive models for EGFR and KRAS mutation status using qualitative radiographic image features. Even though our model outperforms the model created by Gevaert et al. [11] (AUC = 0.89), the present work did not use the same dataset as the first attempt and, consequently, results cannot be directly compared.
The results of this study suggest that an image signature exists that accurately predicts EGFR mutation status but not KRAS. Considering the fact that class proportions are similar in EGFR (26% mutated, 74 % wildtype) and KRAS (25 % mutated, 75% wildtype), the most reasonable hypothesis for this result is that KRAS mutations are not evident through radiographic qualitative features in the same extent as EGFR mutations, which appear to have particular patterns. Gevaert et al. [11], which was also able to create a predictive model for EGFR mutations but not for KRAS, also hypothesised that the results might result from different class proportions between EGFR and KRAS; however in the present study class proportions are similar, which enhances the hypothesis that KRAS mutations do not manifest through qualitative imaging features.
When clinical and imaging data were combined, the performance slightly increased, on average. These results have shown that, although the limited amount of data, images and clinical data combined are potential predictors of EGFR mutation status. However, it is important to further validate the results with a larger dataset, to clarify these features importance.
From the total set of features used in this work, emphysema and lung parenchymal features were the ones that presented a higher correlation with EGFR mutation status.
The model created in this study for the prediction of EGFR mutation status opens interesting opportunities for a better treatment planning and supports oncologists and radiologists with additional information at diagnosis. Key issues to be investigated in the future is whether quantitative features extracted directly from the image have the same predictive ability or even exceed qualitative features.
References
Bakr, S., et al.: A radiogenomic dataset of non-small cell lung cancer. Sci. Data 5, 180202 (2018)
Bakr, S., et al.: Data for NSCLC Radiogenomics Collection (2017). https://doi.org/10.7937/K9/TCIA.2017.7hs46erv. https://wiki.cancerimagingarchive.net/x/W4G1AQ, type: dataset
Breiman, L.: Random forests. Mach. Learn. 45(1), 5–32 (2001)
Chawla, N.V., Bowyer, K.W., Hall, L.O., Kegelmeyer, W.P.: Smote: synthetic minority over-sampling technique. J. Artif. Intell. Res. 16, 321–357 (2002)
Chen, Z., Fillmore, C.M., Hammerman, P.S., Kim, C.F., Wong, K.K.: Non-small-cell lung cancers: a heterogeneous set of diseases. Nat. Rev. Cancer 14(8), 535 (2014)
Clark, K., et al.: The cancer imaging archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging 26(6), 1045–1057 (2013). https://doi.org/10.1007/s10278-013-9622-7
Digumarthy, S.R., Padole, A.M., Gullo, R.L., Sequist, L.V., Kalra, M.K.: Can CT radiomic analysis in NSCLC predict histology and EGFR mutation status? Medicine 98(1) (2019)
Dogan, S., et al.: Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin. Cancer Res. 18(22), 6169–6177 (2012)
Ellison, G., Zhu, G., Moulis, A., Dearden, S., Speake, G., McCormack, R.: EGFR mutation testing in lung cancer: a review of available methods and their use for analysis of tumour tissue and cytology samples. J. Clin. Pathol. 66(2), 79–89 (2013)
Ferlay, J., et al.: Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136(5), E359–E386 (2015)
Gevaert, O., et al.: Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci. Rep. 7, 41674 (2017)
Gevaert, O., et al.: Non–small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data—methods and preliminary results. Radiology 264(2), 387–396 (2012). https://doi.org/10.1148/radiol.12111607, pMID: 22723499
Hames, M.L., Chen, H., Iams, W., Aston, J., Lovly, C.M., Horn, L.: Correlation between KRAS mutation status and response to chemotherapy in patients with advanced non-small cell lung cancer. Lung Cancer 92, 29–34 (2016)
Hansell, D.M., Bankier, A.A., MacMahon, H., McLoud, T.C., Muller, N.L., Remy, J.: Fleischner society: glossary of terms for thoracic imaging. Radiology 246(3), 697–722 (2008)
Janssen-Heijnen, M.L., Coebergh, J.W.W.: Trends in incidence and prognosis of the histological subtypes of lung cancer in North America, Australia, New Zealand and Europe. Lung Cancer 31(2–3), 123–137 (2001)
Liu, Y., et al.: Radiomic features are associated with EGFR mutation status in lung adenocarcinomas. Clin. Lung Cancer 17(5), 441–448 (2016)
Mei, D., Luo, Y., Wang, Y., Gong, J.: CT texture analysis of lung adenocarcinoma: can radiomic features be surrogate biomarkers for EGFR mutation statuses. Cancer Imaging 18(1), 52 (2018)
Mok, T.S., et al.: Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 361(10), 947–957 (2009)
O’dowd, E.L., et al.: What characteristics of primary care and patients are associated with early death in patients with lung cancer in the UK? Thorax 70(2), 161–168 (2015)
Papadopoulou, E., et al.: Determination of EGFR and KRAS mutational status in greek non-small-cell lung cancer patients. Oncol. Lett. 10(4), 2176–2184 (2015)
Riely, G.J., Marks, J., Pao, W.: KRAS mutations in non–small cell lung cancer. Proc. Am. Thorac. Soc. 6(2), 201–205 (2009)
Rosenblatt, F.: The perceptron: a probabilistic model for information storage and organization in the brain. Psychol. Rev. 65(6) (1958)
Scrivener, M., de Jong, E.E., van Timmeren, J.E., Pieters, T., Ghaye, B., Geets, X.: Radiomics applied to lung cancer: a review. Transl. Cancer Res. 5(4), 398–409 (2016)
Siegelin, M.D., Borczuk, A.C.: Epidermal growth factor receptor mutations in lung adenocarcinoma. Lab. Invest. 94(2), 129 (2014)
Varghese, A.M., et al.: Lungs dont forget: comparison of the KRAS and EGFR mutation profile and survival of collegiate smokers and never smokers with advanced lung cancers. J. Thorac. Oncol. 8(1), 123–125 (2013)
Zhou, H., et al.: Poor response to platinum-based chemotherapy is associated with KRAS mutation and concomitant low expression of BRAC1 and TYMS in NSCLC. J. Int. Med. Res. 44(1), 89–98 (2016)
Acknowledgments
This work is financed by the ERDF – European Regional Development Fund through the Operational Programme for Competitiveness and Internationalisation - COMPETE 2020 Programme and by National Funds through the Portuguese funding agency, FCT - Fundação para a Ciência e a Tecnologia within project POCI-01-0145-FEDER-030263.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Dias, C., Pinheiro, G., Cunha, A., Oliveira, H.P. (2019). Radiogenomics: Lung Cancer-Related Genes Mutation Status Prediction. In: Morales, A., Fierrez, J., Sánchez, J., Ribeiro, B. (eds) Pattern Recognition and Image Analysis. IbPRIA 2019. Lecture Notes in Computer Science(), vol 11868. Springer, Cham. https://doi.org/10.1007/978-3-030-31321-0_29
Download citation
DOI: https://doi.org/10.1007/978-3-030-31321-0_29
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-31320-3
Online ISBN: 978-3-030-31321-0
eBook Packages: Computer ScienceComputer Science (R0)