Abstract
To interpret a breast MRI study, a radiologist has to examine over 1000 images, and integrate spatial and temporal information from multiple sequences. The automated detection and classification of suspicious lesions can help reduce the workload and improve accuracy. We describe a hybrid mass-detection algorithm that combines unsupervised candidate detection with deep learning-based classification. The detection algorithm first identifies image-salient regions, as well as regions that are cross-salient with respect to the contralateral breast image. We then use a convolutional neural network (CNN) to classify the detected candidates into true-positive and false-positive masses. The network uses a novel multi-channel image representation; this representation encompasses information from the anatomical and kinetic image features, as well as saliency maps. We evaluated our algorithm on a dataset of MRI studies from 171 patients, with 1957 annotated slices of malignant (59%) and benign (41%) masses. Unsupervised saliency-based detection provided a sensitivity of 0.96 with 9.7 false-positive detections per slice. Combined with CNN classification, the number of false positive detections dropped to 0.7 per slice, with 0.85 sensitivity. The multi-channel representation achieved higher classification performance compared to single-channel images. The combination of domain-specific unsupervised methods and general-purpose supervised learning offers advantages for medical imaging applications, and may improve the ability of automated algorithms to assist radiologists.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others

Keywords
1 Introduction
Magnetic Resonance Imaging (MRI) of the breast is widely-used as a screening examination for women at high risk of breast cancer. A typical breast MRI study consists of 1000 to 1500 images, which involve a lot of time for interpretation and reporting. Computer-assisted interpretation can potentially reduce the radiologist’s workload by automating some of the diagnostic tasks, such as lesion detection. Previous studies addressed automatic lesion detection in breast MRI using a variety of methods. These can be broadly categorized into: image processing approaches [1,2,3], machine learning approaches [4, 5], or a combination of both [6,7,8]. Deep convolutional neural networks (CNN) have been previously applied to breast MRI images for mass classification [9], as well as parts of an automated lesion segmentation pipeline [10]. Cross-saliency analysis has been shown to be advantageous for unsupervised asymmetry detection, and was applied to lesion detection in breast mammograms and brain MRI [11].
We describe a new hybrid framework for the automatic detection of breast lesions in dynamic contrast enhanced (DCE) MRI studies. The framework combines unsupervised candidate proposals by analyzing salient image areas, with a CNN classifier that filters out false detections using multiple image channels. Our work comprises four major contributions: (1) an unsupervised lesion detection algorithm, using patch-based distinctiveness within the entire breast, and between the left and right breasts; (2) a new multi-channel representation for DCE MRI images, which compactly captures anatomy, kinetic, and salient features in a single image that can be fed to a deep neural network; (3) a hybrid lesion detection framework that provides high detection accuracy and a low false positive rate by combining the unsupervised detection and the multi-channel representation with a deep neural network classifier; (4) the evaluation of proposed methods on a large dataset of MRI studies, including publicly available data.
2 Methods
2.1 Datasets
To train and evaluate our system, we used a dataset of 193 breast MRI studies from 171 female patients, acquired through a variety of acquisition devices and protocols. Two publicly available resources [12,13,14] provided the data of 78 patients (46%). Each study included axial DCE T1 sequences with one pre-contrast series and at least 3 post-contrast series. Three breast radiologists interpreted the studies using image information alone, without considering any clinical data. Each identified lesion was assigned a BI-RADS score. The boundaries of the lesions were manually delineated on each relevant slice, with an average of 11 ± 10 annotated slices per patient. Overall, there were 1957 annotated lesion contours in 1845 slices; 59% of them were labeled as malignant (BI-RADS 4/5) and 41% were labeled as benign (BI-RADS 2/3). The average lesion size was 319 ± 594 mm2. We partitioned the patients into training (75%, 128 patients, 1326 slices) and testing (25%, 43 patients, 519 slices) subsets. The partitioning was random, while ensuring a similar distribution of benign and malignant lesions in each of the subsets.
2.2 Image Representation
We used our multi-channel image representation previously described in [9]. The rationale of this representation is to capture both anatomical and metabolic characteristics of the lesion in a single multi-channel image. Figure 1 shows the three image channels that represent the DCE study: (1) peak enhancement intensity channel; (2) contrast uptake channel: difference between peak enhancement and baseline images; (3) contrast washout image: difference between the early and the delayed contrast images.
Axial DCE T1 sequences acquired before contrast injection (a), at peak enhancement (b) and after contrast washout (c). The kinetic graph (d) shows the pattern of contrast uptake and the temporal location of each sequence. The multi-channel image representation combines the peak-enhancement image (b) with the contrast uptake image (e) and contrast washout image (f).
2.3 Lesion Detection Framework
Figure 2 shows the components of the lesion detection framework.
Image Preprocessing and Segmentation
The two-dimensional slice images were normalized to reduce data variability due to the mixture of studies from different sources. For each of the data subsets, the global 1% and 99% percentiles of pixel intensity were calculated for each channel, and contrast stretching was applied to convert all images to the same dynamic range. The breast area was segmented using U-Net, a fully convolutional network designed for medical image segmentation [15]. The network was implemented using Lasagne, a python framework built on top of Theano. We trained the network on a subset of slices with manually delineated breast contours. The training process of 20 epochs on a batch size of 4 images required about 1 h on a Titan X NVIDIA GPU. The remainder of the detection pipeline was applied only to the region within the segmented breast.
Saliency Analysis
For each MRI slice image, two patch-based saliency maps were created [11]: (1) patch distinctiveness and, (2) contralateral breast patch flow. The patch distinctiveness saliency map is generated by computing the L1-distance between each patch and the average patch along the principal components of all image patches [16]. For a given vectorized patch px,y around the points (x, y), this measure is given by:
where \( \omega_{k}^{T} \) is the kth principal component of the entire image patch distribution.
Contralateral patch flow calculates the flow field between patches of the left and right breasts, using the PatchMatch algorithm [17]. The algorithm uses the smooth motion field assumption to compute a dense flow field for each pixel by considering a k x k patch around it. Initially, for each pixel location (x, y), it assigns a random displacement vector T that marks the location of its corresponding patch in the other image. The quality of the displacement T is then measured by computing the L2 distance between the corresponding patches:
The algorithm attempts to improve the displacement for each location by testing new hypotheses generated from the displacement vectors of neighboring patches in the same image. The algorithm progresses by iterating between the random and propagation steps, keeping the location of the best estimate according to the L2 distance. We applied the algorithm to find, for each patch in the source image, the corresponding nearest neighbor patch in the target image. The nearest neighbor error (NNE) was used to estimate the cross-image distinctiveness of the patch:
Candidate Region Detection
Candidate regions were detected on the saliency maps using a scale-invariant algorithm that searches for regions with high density of saliency values. For a given range of window sizes (wi, hj) and a set of threshold values {t1, t2,.. tn}, the algorithm efficiently computes, for each pixel (x, y) and a region \( s_{x,y} \) of size wi x hj around it:
Non-maximal suppression was then applied to the Score image to obtain the locations of all local maxima. We used window sizes in the range of 5 to 50 pixels and normalized threshold values from 0.3 to 0.9. The region detection algorithm was applied to each of the two saliency maps, producing two binary detection masks, which were combined by an ‘or’ operator to generate candidate detections per slice.
CNN-Based Candidate Classification
The detected candidates were cropped from their slice images using square bounding boxes, extended by 20% to ensure that the entire lesion was included in the cropped image. The extracted lesion images were resized to fit the CNN input, and 32 × 32 × 5 multi-channel images were created with 3 channels of the DCE image and 2 channels of the corresponding saliency maps. The CNN architecture consisted of 9 convolutional layers in 3 consecutive blocks, similar to [18]. The first block had two 5 × 5 × 32 filters with ReLU layers followed by a max pooling layer, the second block had four 5 × 5 × 32 filters with ReLU layers followed by an average pooling layer, and the final block had three convolutional layers of size 5 × 5 × 64, 6 × 6 × 64, and 3 × 3 × 64 respectively, each followed by a ReLU layer. The network was terminated by a fully connected layer with 128 neurons and a softmax loss layer. The network output assigned either a ‘mass’ or ‘non-mass’ label to each bounding box. As the training data was unbalanced, with many more examples of ‘non-mass’ regions, we trained an ensemble of 10 networks, each with a different random sample of ‘non-mass’ regions. Majority voting of the ensemble determined the final classification.
For each slice, the output of the framework was a binary detection map, with regions that were proposed by the saliency analysis and classified as ‘mass’ by the CNNs. The detection output per study was generated by summing the slice detection maps along the longitudinal axis. This produced a projected heatmap showing the spatial concentration of detected regions. Thresholding this heatmap was used to further reject false detections.
2.4 Experiments
We trained the ensemble of convolutional networks on a set of 1564 bounding boxes of masses and 11,286 of non-masses, detected by the saliency analysis. The training set was augmented by adding three rotated and two flipped variants for each image. The networks were trained using MatCovNet, using a stochastic gradient descent solver with a momentum of 0.9. The average training time of 100 epochs was 20 min on NVIDIA-Titan-X black GPU. To evaluate the performance of the detection framework on the test set of 43 patients, all DCE slices of the test studies were processed by the entire pipeline. Overall, there were 5420 test slices, an average of 126 slices per patient. We compared the detection maps per slice and per study to the annotated ground-truth.
3 Results
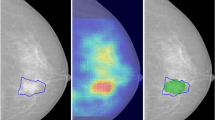
The unsupervised saliency analysis correctly detected 0.96 of true lesions in the entire dataset, with an average of 9.7 false positive detections per slice. The detection rates on the training and testing sets were similar. The average accuracy of mass/non-mass classification, obtained by the CNN on the validation set during the training process, was 0.86 ± 0.02, with area under the receiver operator characteristics curve (AUC) of 0.94 ± 0.01. Table 1 shows that training with the 5-channel image representation achieved the highest accuracy. The evaluation of the entire detection framework on the test set slices yielded a sensitivity of 0.85 with 0.7 false-positives per slice. The CNN was able to reject 89% of the false candidate regions detected by the saliency analysis. Comparing the detection heatmaps per study with the projected ground-truth showed an improved sensitivity of 0.98 with an average of 7 false-positive detections per study (Table 2). Figure 3 shows a representative example of true-positive and false-negative detections.
An example of true-positive and false-negative detection. A BI-RADS 5 invasive ductal carcinoma lesion in the right breast, shown in two peak-enhancement slices of the same sequence (a, c), along with the corresponding cross-saliency maps (b, d) and the ground-truth contour (red). In (b), the detection algorithm identified the region of the lesion, and correctly classified it (green), while rejecting false detections (yellow). The same lesion at a consecutive slice was missed by the saliency analysis (d).
4 Discussion
Despite the potential of computer-assisted algorithms to improve the reading process of breast imaging studies, and although existing systems have been shown to improve sensitivity, they have not significantly affected the diagnostic accuracy (AUC) or the interpretation times of both novice and experienced radiologists [19]. Recent advancements in deep learning technologies provide an opportunity to develop learning-based solutions that will effectively support the diagnostic process. However, as most deep learning research has focused on natural images, using deep networks as a ‘black-box’ solution for detection and classification problems in the medical imaging domain may not provide optimal results. Our suggested hybrid approach, which combines tailored computer vision algorithms of cross-saliency analysis with deep network classifiers, allows the incorporation of domain-specific insights about the data to improve performance. In our framework, both the multi-channel representation of DCE images and the cross-saliency analysis of left and right breasts are examples of such insights. The great majority of published work on lesion detection in breast MRI uses proprietary datasets, typically small in size, which could not be used as a common benchmark for comparison. The reported results show a large variability in sensitivity and false positive rate, ranging from 1 to 26 false positives per study at a sensitivity range of 0.89 to 1.0 [1, 2, 10]. An objective performance comparison between methods requires publicly available large datasets with ground-truth annotations. In this work, we used MRI studies from the TCIA repository [12], enriched by additional proprietary studies. Additionally, sharing domain-specific deep learning models that have been trained on proprietary data may be an alternative mechanism for collaboration among medical imaging researchers. Our current work is missing a comparison to state-of-the-art detection methods such as faster R-CNN, which provides both region proposals and classification within the same convolutional network. The plans for our future work include such a comparison to test our hypothesis on the advantages of the hybrid approach.
In conclusion, we propose a combination of multi-channel image representation, unsupervised candidate proposals by saliency analysis, and deep network classification to provide automatic lesion detection in breast MRI with high sensitivity and low false-positive rate. When evaluated on a large set of studies, this method could facilitate the incorporation of cognitive technologies into the radiology workflow.
References
Vignati, A., Giannini, V., et al.: A fully automatic lesion detection method for DCE-MRI fat-suppressed breast images, 26 February 2009
Ertas, G., Doran, S., Leach, M.O.: Computerized detection of breast lesions in multi-centre and multi-instrument DCE-MR data using 3D principal component maps and template matching. Phys. Med. Biol. 56, 7795–7811 (2011)
McClymont, D., Mehnert, A., et al.: Fully automatic lesion segmentation in breast MRI using mean-shift and graph-cuts on a region adjacency graph. J. Magn. Reson. Imaging 39, 795–804 (2014)
Gallego-Ortiz, C., Martel, A.L.: Improving the accuracy of computer-aided diagnosis for breast MR imaging by differentiating between mass and nonmass lesions. Radiology 278, 679–688 (2016)
Agner, S.C., Soman, S., et al.: Textural kinetics: a novel dynamic contrast-enhanced (DCE)-MRI feature for breast lesion classification. J. Digit. Imaging 24, 446–463 (2011)
Ertaş, G., Gülçür, H.Ö., et al.: Breast MR segmentation and lesion detection with cellular neural networks and 3D template matching. Comput. Biol. Med. 38, 116–126 (2008)
Renz, D.M., Böttcher, J., et al.: Detection and classification of contrast-enhancing masses by a fully automatic computer-assisted diagnosis system for breast MRI. J. Magn. Reson. Imaging 35, 1077–1088 (2012)
Pang, Z., Zhu, D., Chen, D., Li, L., Shao, Y.: A computer-aided diagnosis system for dynamic contrast-enhanced MR images based on level set segmentation and ReliefF feature selection. Comput. Math. Methods Med. 2015(2015), Article ID 450531 (2015). doi:10.1155/2015/450531
Amit, G., Ben-Ari, R., Hadad, O., Monovich, E., Granot, N., Hashoul, S.: Classification of breast MRI lesions using small-size training sets: comparison of deep learning approaches. In: Proceedings of SPIE Medical Imaging, vol. 10134 (2017)
Wu, H., Cristina Gallego-Ortiz, A.M.: Deep artificial neural network approach to automated lesion segmentation in breast DCE-MRI. In: Proceedings of the 3rd MICCAI Workshop on Breast Image Analysis, pp. 73–80 (2015)
Erihov, M., Alpert, S., Kisilev, P., Hashoul, S.: A cross saliency approach to asymmetry-based tumor detection (2015)
Clark, K., Vendt, B., et al.: The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging. 26, 1045–1057 (2013)
Lingle, W., Erickson, B.J., et al.: Radiology Data from The Cancer Genome Atlas Breast Invasive Carcinoma [TCGA-BRCA] collection. http://doi.org/10.7937/K9/TCIA.2016.AB2NAZRP
Bloch, B.N., Jain, A., Jaffe, C.C.: Data from breast-diagnosis. http://doi.org/10.7937/K9/TCIA.2015.SDNRQXXR
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). doi:10.1007/978-3-319-24574-4_28
Margolin, R., Tal, A., Zelnik-Manor, L.: What makes a patch distinct? In: 2013 IEEE Conference on Computer Vision and Pattern Recognition, pp. 1139–1146. IEEE (2013)
Barnes, C., Shechtman, E., Goldman, D.B., Finkelstein, A.: The generalized PatchMatch correspondence algorithm. In: Daniilidis, K., Maragos, P., Paragios, N. (eds.) ECCV 2010. LNCS, vol. 6313, pp. 29–43. Springer, Heidelberg (2010). doi:10.1007/978-3-642-15558-1_3
Hadad, O., Bakalo, R., Ben-Ar, R., Hashoul, S., Amit, G.: Classification of breast lesions using cross-modal deep learning. In: IEEE International Symposium on Biomedical Imaging (ISBI) (2017)
Lehman, C.D., Blume, J.D., et al.: Accuracy and interpretation time of computer-aided detection among novice and experienced breast MRI readers. Am. J. Roentgenol. 200, W683–W689 (2013)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Amit, G. et al. (2017). Hybrid Mass Detection in Breast MRI Combining Unsupervised Saliency Analysis and Deep Learning. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds) Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science(), vol 10435. Springer, Cham. https://doi.org/10.1007/978-3-319-66179-7_68
Download citation
DOI: https://doi.org/10.1007/978-3-319-66179-7_68
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-66178-0
Online ISBN: 978-3-319-66179-7
eBook Packages: Computer ScienceComputer Science (R0)