Abstract
Most past research on sequential sampling models of decision-making have assumed a time homogeneous process (i.e., parameters such as drift rates and boundaries are constant and do not change during the deliberation process). This has largely been due to the theoretical difficulty in testing and fitting more complex models. In recent years, the development of simulation-based modeling approaches matched with Bayesian fitting methodologies has opened the possibility of developing more complex models such as those with time-varying properties. In the present work, we discuss a piecewise variant of the well-studied diffusion decision model (termed pDDM) that allows evidence accumulation rates to change during the deliberation process. Given the complex, time-varying nature of this model, standard Bayesian parameter estimation methodologies cannot be used to fit the model. To overcome this, we apply a recently developed simulation-based, hierarchal Bayesian methodology called the probability density approximation (PDA) method. We provide an analysis of this methodology and present results of parameter recovery experiments to demonstrate the strengths and limitations of this approach. With those established, we fit pDDM to data from a perceptual experiment where information changes during the course of trials. This extensible modeling platform opens the possibility of applying sequential sampling models to a range of complex non-stationary decision tasks.
Similar content being viewed by others
Introduction
Most past decision research has been devoted to understanding simple, two-choice decisions where a choice is made on the basis of fixed, unchanging information, or on information that changes randomly around a fixed central tendency (Ratcliff 1978; Busemeyer and Townsend 1993; Shadlen and Newsome 1996; Gold and Shadlen 2001; Smith and Ratcliff 2004). Sequential sampling models such as the diffusion decision model (DDM) have been widely used in both cognitive psychology and applied domains to account for these types of simple decisions (Ratcliff, 1978; Smith & Ratcliff, 2004, Ratcliff, Thapar, & McKoon, 2001; White, Ratcliff, Vasey, & McKoon, 2010). While Ratcliff (1980) first discussed the importance of examining more complex decision tasks involving changing information, modeling the impact of non-stationary inputs on decision-making has traditionally received less attention, but recently there has been increasing interest from a number of perspectives. These include dynamic environments across trials (Bröder & Schiffer, 2006; Gureckis & Love, 2009), non-stationary decision criteria (Verbruggen & Logan 2008; Logan & Burkell, 1986), and changed information due to internal factors (e.g., eye movements or attention switching) (Diederich, 1997; Diederich & Busemeyer, 1999; Krajbich, Armel, & Rangel, 2010; Krajbich & Rangel 2011).
One of the emerging approaches to modeling non-stationary decision tasks involves extending standard sequential sampling models such as DDM and the linear ballistic accumulator (LBA) model to have piecewise constant, time-varying properties (drift rates for example). These “piecewise” models have been used to explain a range of phenomena including the relationship between eye movements and choice behavior (Krajbich et al. 2010; Krajbich and Rangel 2011), multi-attribute decision-making (Diederich 1997; Diederich and Oswald 2016), the interplay between multiple thinking modes during decision-making (Guo, Trueblood, & Diederich, 2015; Diederich & Trueblood, submitted), and the impact of exogenous changes of information during decision-making (Holmes, Trueblood, & Heathcote, 2016) such as brief motion pulses in random moving dot displays (Tsetsos, Gao, McClelland, & Usher, 2012) and fluctuations of brightness in simple perceptual displays (Tsetsos, Usher, & McClelland, 2011). Despite the growing interest in modeling complex decision tasks with non-stationary inputs, this approach has been limited for mathematical and computational reasons.
There has been significant research on developing methods to implement DDM (Voss & Voss,2007; Vandekerckhove & Tuerlinckx, 2007; 2008), including hierarchical Bayesian methods (Vandekerckhove, Tuerlinckx, & Lee, 2011; Wiecki, Sofer, & Frank, 2013; Wabersich & Vandekerckhove, 2014). In particular, past work has lead to the development of quick and efficient methods for implementing diffusion models (Navarro and Fuss 2009), which have been used in applications such as the neural drift diffusion model (Turner, Van Maanen, & Forstmann, 2015). However, there has been much less research on methods for implementing piecewise variants of this model (which we call the piecewise diffusion decision model or pDDM). In particular, most quantitative fitting of pDDM utilizes maximum likelihood methods to fit mean choice and response time data (see for example, Krajbich et al. 2010). However, simply fitting mean response time data is not ideal and can lead to drastically different parameter estimates than fits to the full distribution (Turner and Sederberg 2014).
The goal of the present paper is to present hierarchical Bayesian methods for implementing pDDM and to evaluate the efficacy of those methods. The difficulty in implementing pDDM in a hierarchical Bayesian method is that doing so requires the ability to calculate model likelihoods up to a billion times or more. While methods (involving series solutions) for calculating these likelihoods have been devised for single (Navarro and Fuss 2009) and multi-stage (Srivastava, Feng, Cohen, Leonard, & Shenhav, 2017) models, it is not clear if these methods are sufficiently efficient for this application. Here, we describe how simulation along with a recently developed approximate Bayesian method, called the probability density approximation (PDA) method (Turner and Sederberg 2014; Holmes 2015), can be used to accomplish this. This method is an alternative to existing Bayesian and approximate Bayesian computation (ABC) parameter estimation methodologies that do not require either an analytically tractable model likelihood function or the use of sufficient statistics for the parameters of interest (which is needed for ABC). This method has previously been used to fit other sequential sampling models such as the leaky competing accumulator model (Turner et al. 2014) and the piecewise linear ballistic accumulator (pLBA) model (Holmes 2015; Holmes et al. 2016). However, it has not yet been evaluated in the context of pDDM.
We begin by reviewing the basic idea underlying the PDA method. In order to demonstrate this method in a simpler context, we next apply this method to the standard DDM (no changes of information). Next, parameter recovery is performed for pDDM to check the effectiveness of this method at estimating parameters of a time-heterogenous process. To demonstrate the application of this modeling in estimating the influence of changing information on decisions, model inferences associated with a new experiment where information changes during the course of a decision are presented. The final section (prior to the discussion) provides additional technical details, including a discussion of how estimation errors influence likelihood approximation and timing data. Data for the flashing grid experiment discussed below along with parameter recovery codes for the DDM and pDDM models are provided on the Open Science Framework (osf.io/dkmbk/).
The PDA method
Here, we will discuss the relevant details for applying the PDA method. Further information and details can be found in Holmes (2015) and Turner and Sederberg (2014). For the remainder of this paper, the term “data” will refer to choice/response time (RT) data, which we denote as X, though the method can be applied to any data set that continuously varies with respect to a single axis (time in this case).
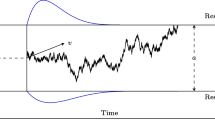
The central task of Bayesian parameter estimation is to compute the posterior density function for a model π(𝜃|X), which provides a principled way to determine for which parameter combinations the model best accounts for the observed data. Markov chain Monte Carlo methods provide a powerful framework for approximating this density function. However, they require the user to provide an analytically tractable likelihood function L(𝜃|X). Unfortunately, many models, particularly models involving temporal changes of the decision process as considered here, do not emit an analytically tractable likelihood function (Fig. 1).
pDDM schematic representation. As with the standard DDM, the pDDM represents accumulation of evidence over time with parameters representing rates of accumulation (μ), strength of stochasticity (s), start point bias (z), response thresholds (at 0 and a), and non-decision time (t N D ). To account for changes of information, pDDM allows for evidence-accumulation rates to change at a discrete point in time. To account for any lag between presentation and accumulation of new information, a delay parameter t d e l a y is also included. In addition to these parameters, variance parameters representing across trial variability in start point (s z ) and accumulation rate (s μ ) are also included
The PDA method, which we use here, is a simulation-based method that utilizes tools from non-parametric statistics to overcome the lack of a tractable likelihood function. This method uses a standard Metropolis Hastings (MH) framework to generate MCMC proposals and accept or reject them on the basis of quality of fit. The key step in this process is the computation of the likelihood L(𝜃|X) of a proposed parameter set, which is the measure of quality of fit used to accept or reject it. Assuming each trial is independent (which we will assume from here on), the full likelihood can be decomposed into the product of likelihood of individual observations
The key to the PDA is the construction of an approximation of the likelihood of each individual observation \(\hat {L} (\theta | x_{i})\) , which can be directly substituted into the MH algorithm. From here on, \(\hat {~}\) will denote an approximation; in this case it denotes the approximation of L(𝜃|x i ). The first step in this construction is the simulation of a synthetic data set \(\tilde {X}\) consisting of N s simulations from the model with the proposed parameter set 𝜃. In the case of stochastic process model such as DDM, this requires the simulation of stochastic differential equations describing evidence accumulation to generate synthetic choice response time data. The second step is to use this data to construct a density function representing this simulated response time data. The kernel density estimate (KDE), commonly used in non-parametric statistics, is a powerful tool for doing just this Silverman (1982) and Silverman (1986).
The basic density estimation problem is to determine the likelihood \(\hat {L} (\theta | x)\) at a point x from a collection of simulated model observations \(\tilde {X}=\{\tilde {x}_{j}\}\), where j = 1...N s . This density value can be approximated using the KDE as
Here K h is a “smoothing kernel” defined by
where K is a continuous function that is symmetric about zero and integrates to 1. The parameter h is a “bandwidth” (akin to the bin size in a histogram) and determines the smoothing properties of the kernel: larger h yields more smoothing of the distribution. Using this method, an estimate for each \(\hat {L} (\theta | x_{i})\) can be computed and substituted into Eq. (1) to compute a final likelihood, which is subsequently used by the MH algorithm to choose whether to accept or reject 𝜃. Figure 2 demonstrates the construction of the approximate likelihood function \(\hat {L} (\theta | x)\) for the DDM model. As this figure demonstrates, this method can generate high-quality density function approximations using a relatively simple and well-established technique.
Likelihood construction. To demonstrate the effectiveness of PDA likelihood construction, a data set comprised of 1000 sample choice/RTs is generated to mimic a participant data set. Model parameters used to generate this data set are a = 0.08,z = 0.04,s z = 0.05,μ = 0.05,s μ = 0.05,t n d = 0.15, and s = 0.05. A simulation timestep of d t = 0.0001 was used to stochastically simulate this data set. The circles show the choice/RT distribution for this data. Subsequently, a synthetic likelihood is constructed using the PDA method with N s = 10,000,h = 0.02, and d t = 0.01 seconds. Black and gray indicate the probability of correct and incorrect responses, respectively
Accounting for continuous response times and discrete choices
In response-time models such as DDM, there are usually two types of response data available, time and choice. Thus, each response observation (utilizing the notation introduced above) would take the form x i = (t i , c i ) where t i is the observed response time and c i the choice. The PDA discussion above centered around how to approximate the likelihood of a continuous variable. In the context of this paper, that continuous variable is the response time. Choice responses, which are discrete, however, should not be neglected. The common way this data is handled (as is done here) is to break the data set into a collection of defective probability distributions, each associated with a particular choice alternative. In the case of two- choice alternatives, this would yield two defective densities whose total probability mass sum to 1. The PDA method is used to construct each of these densities separately. For details of the minor complexities associated with ensuring that the PDA produces a defective distribution and that the resulting approximate distributions mass sum to 1, see Turner and Sederberg (2014) and Holmes (2015).
DDM parameter recovery
In subsequent sections, we will use this method to fit a piecewise variant of the canonical DDM model with piecewise constant, time-varying drift rates. In order to demonstrate and assess this method in a more simplified context, however, we will consider the canonical DDM model first. Here, we perform a parameter recovery experiment for the DDM model parameterized by a start point bias (z), trial-to-trial variability in that bias (s z ), evidence threshold (a), drift rate (μ), trial-to-trial variability in drift rate (s μ ), non-decision time (t N D ), and moment-to-moment accumulation stochasticity (s). To constrain the model and remove the scaling degeneracy in this model, we fix s = 0.05 so that it has a similar size as the parameters used for simulation study (see Fig. 2 caption).
The DDM model will be fit to a synthetic, baseline data set consisting of 2000 simulated observations (see the Technical Methods section for further details). To perform this parameter estimation, we use a differential evolution MCMC (DEMCMC) (Ter Braak, 2006; Turner, Sederberg, Brown, & Steyvers, 2013) procedure to fit parameters with the following uninformative priors
where U denotes the uniform distribution on the specified interval. The methodological parameters (N s , h, d t) are those chosen from the analysis in the Technical Methods section. As was performed in Holmes (2015), a likelihood resampling procedure is implemented (resampling every four chain iterations) to avoid stuck chains and improve chain acceptance rates, see Holmes (2015) for further details on resampling.
Figure 3a shows the posterior distribution for all parameters along with the exact parameter values (grey bars) used to generate the data set. Results indicate that this method performs well at recovering the parameters a, z, μ, t N D , which are those primarily used to draw psychological inferences. The posteriors for trial-to-trial variability parameters are less constrained and in particular s μ does not appear to fit well. Trial-to-trial variability parameters are, however, rarely used (to our knowledge) for inferences and estimating them is a challenge for any method.Footnote 1 The prior distributions were chosen to be as uninformative as possible, being two orders of magnitude larger than the parameters themselves. A tenfold reduction in the scale of these priors has little effect on fits, however, and it appears the results are not highly sensitive to the prior.
DDM parameter recovery. a Posterior distributions for all parameters of the DDM model. A data set consisting of 2000 trials was generated using the same parameters as in Fig. 2. The stochastic parameter s = 0.05 is fixed and not estimated. Gray lines show exact values for parameters while dark curves show the posterior distributions. b Quality of model fit. The model fit to data is shown for both the mean value of the posterior parameter distribution (solid line) and the exact parameter set (dashed line), with the RT distribution of the underlying data indicated with dots
It is important to note that parameter recovery here is not perfect, though the exact parameter values used to generate the simulated data are well within the credible interval for each parameter. This, however, is a feature of the DDM model itself rather than the fitting method. To illustrate this, Fig. 3b compares the RT distribution of the data itself to the RT distribution computed from the exact and best fit (mean of the posterior) parameter values. Results show that the distributions generated by best fit and exact parameters are nearly indistinguishable. This is a common phenomenon in modeling literature refereed to as parameter “sloppiness” (Apgar, Witmer, White, & Tidor, 2010; Gutenkunst et al., 2007), which describes models whose predictions are relatively insensitive to the precise values of parameters (see General Discussion for further explanation). Brown, Ratcliff, and Smith (2006) noted a similar phenomenon referred to as parameter “inflation” where numerical approximation errors arising from the simulation of stochastic differential equations led to imperfect parameter recovery. They however also noted that this inflation did not influence goodness of fit.
We note that this is but one example demonstrating the ability of PDA to recover parameters. Similar studies have shown that it is capable of recovering parameters of a bimodal distribution (Holmes 2015) as well as the parameters of the linear ballistic accumulator (LBA) model (Turner and Sederberg 2014; Holmes 2015). This method was shown to adequately recover parameters of a piecewise variant of the LBA model (pLBA) (Holmes et al. 2016; Holmes 2015), though there were some complexities associated with the “sloppiness” of that model as well that were independent of the PDA method. It was also shown to successfully recover the parameters of a hierarchal model (Turner and Sederberg 2014). While we take this collection of studies as evidence of the efficacy of this method, it should still be used with caution. While in the limit (N s →∞, h → 0) this method is guaranteed to exactly reproduce model likelihoods, in practice its approximate nature will always introduce some errors that will influence the fitting process. For further discussion, see Holmes (2015).
pDDM parameter recovery
Next, we assess the effectiveness of this model at fitting the pDDM model. This is an extension of the classical DDM model to account for a change of information during the course of a trial. In Holmes et al. (2016) for example, participants performed a random dot motion (RDM) task where the motion of dots changed once during each trial. Here we consider a simple extension of the DDM where only a single change of information, which is associated with a change of drift rate, occurs (Fig. 1). This model is described by a start point bias (z), trial-to-trial variability in that bias (s z ), evidence threshold (a), drift rate (μ b e f o r e ) before the change of information, drift rate (μ a f t e r ) after the change of information, trial-to-trial variability in these drift rates (s μ ) (assumed to be the same for μ b e f o r e and μ a f t e r ), a delay parameter (t d e l a y ) accounting for a potential delay between the presentation and integration of new information, non-decision time (t N D ), and moment to moment accumulation stochasticity (s). As before, (s) will always be fixed to constrain the model. For simplicity, we do not consider here trial-to-trial variability in non-decision time, though it could be included.
We begin by first generating a baseline data set consisting of 1000 model simulations with no change of information and an additional 1000 model simulations with a change of information mid trial, for a total of 2000 observations. For ‘switch trials’ where there is a change of information during the course of a trial (e.g., direction of dot motion in the RDM task), this change was coded to occur at T s = 0.5 s. We note however that a simulated decision is allowed to occur prior to T s on these trials, as would be the case for a participant responding quickly in an experiment. This structure mimics the RDM task used in Holmes et al. (2016). To generate this data set, we again used an Euler Maruyama (EM) scheme to simulate the drift diffusion SDE with d t = 0.0001. The values of the parameters used to generate this data set are in the caption to Fig. 4. The same DEMCMC procedure was used to fit this model with the following uninformative priors (which again do not strongly influence results)
Results (Fig. 4) show that as with the DDM model, the PDA is effective at estimating the values of a, z, t d e l a y , μ b e f o r e , μ a f t e r , though it performs poorly at estimating the trial to trial variances s μ , s z . Finally, we note that the posteriors for μ a f t e r and t d e l a y are not as well localized as for a, z, t N D , μ b e f o r e . This was found for a piecewise extension of the LBA model (pLBA) as well (Holmes et al. 2016).
pDDM parameter recovery. a Posterior distributions for all parameters of the pDDM model. A data set consisting of 1000 non-switch and 1000 switch trials was generated using the following parameters: a = 0.08,z = 0.04,s z = 0.01,μ b e f o r e = 0.05,μ a f t e r = −0.05,s μ = 0.05,t N D = 0.15,t d e l a y = 0.25. For simulated switch trials, the switch was coded to occur at T s = 0.5 s. The stochastic parameter s = 0.05 is fixed and non-estimated. Gray lines show exact values for parameters while dark curves show the posterior distributions. b The same as (a) but with 5000 switch and non-switch simulated observations each
This widely spread posterior for a subset of parameters could result from failures of the method or highly correlated model parameters. One common way to look for parameter correlations is to calculate or plot all pairwise correlations among parameters from the saved MCMC chain data. Another, which we utilized here, is to perform a principal component analysis (PCA) of the MCMC chain data. This has both advantages and disadvantages over the correlation plot approach. The disadvantage is that PCA is only effective at detecting linear correlations. The advantages are that (1) it detects high dimensional rather than pairwise correlations and provides information about which parameters are involved and (2) it provides a measure of how much of the variability of the posterior comes from each correlation. Applying a PCA to the MCMC chain data here, we find that a single principle component is responsible for ∼ 98% of the chain variance. Inspection of the PCA eigenvectors shows that the principle component nearly aligns with the t d e l a y axis. Thus, a vast majority of the chain variability results from imprecision in the estimate of the delay parameter, and this is an artifact of the model more so than the method. Despite this, the peak of the t d e l a y parameter coincides with the known value. For further discussion of this PCA approach with a more in-depth example, see Holmes (2015).
Given that only switch trials where responses occur after the T s + T d e l a y inform the T d e l a y and μ a f t e r parameters, we increased the size of the data set to 5,000 switch and non-switch trials, respectively. Results show that posteriors are slightly more localized, but only marginally. Thus, simply adding more data of the same type does not yield a discernible benefit from a theoretical perspective.
pDDM fitting of flashing grid data
Next, we apply the PDA to fit the pDDM model to an experimental data set. Our focus here is not to draw psychological conclusions. Rather, it is to demonstrate the use of modeling methodology. We first describe the experimental protocol.
Materials and methods
Participants
In total, 18 Vanderbilt University students completed the study for course credit. All participants gave informed consent to participate. Three participants were excluded from the modeling because of low accuracy after the initial practice blocks. These participants were most likely not engaged throughout the task.
Procedure
Stimuli were ‘flashing grids’, a square grid filled with two different colored elements (see example in Fig. 5). The two colors were blue and pink. The grid flickered so that every three frames the elements were randomly rearranged. Each element in the grid was a 10 ×10 pixel square. The grid had 20 elements in each row and 20 elements in each column for a total of 400 colored elements. The grids were presented on a black background and displayed on a 60-Hz monitor. In some of the trials, the proportion of blue to pink elements remained constant throughout the trial. In the remaining trials, the proportion of blue to pink elements changed once midway through the trial. On these ‘switch’ trials, the proportion of elements was simply reversed (e.g., if 60% of the elements were blue at the start of the trial, then it switched to 60% pink).
Flashing grid stimulus. Each grid was filled with elements of two colors (blue and pink). The location of the elements was randomly rearranged every three frames. Participants were asked to decide if the grid had more blue or pink elements. On some trials, the proportion of colored elements changed midway through the trial
At the beginning of the experiment, participants were instructed that they would see a flashing grid filled with blue and pink elements and their task was to decide if there are more blue or pink elements in the grid. They were told “you will enter your choices by pressing the ‘z’ key if you think the grid has more pink elements and ‘m’ key if you think the grid has more blue elements.” Participants were asked to place the index finger of their left hand of the ‘z’ key and the index finger of their right hand on the ‘m’ key throughout the experiment. They were also told that they would complete many blocks of trials, that some trials would be harder than others, and that they would sometimes receive feedback about their responses. Participants were asked to respond to the best of their ability as quickly as possible, but were not told that the proportion of colored elements could possibly change during some trials.
At the beginning of each trial, they viewed a fixation cross for 250 ms, then there was a blank screen for 100 ms, followed by the stimulus. Participants had up to 2 s to view the stimulus and give a response or the trial terminated by itself and a non-response was recorded. Otherwise, the trial terminated immediately after a response was made. The fixation cross for the next trial appeared immediately after the termination of the previous trial.
Participants completed 24 blocks of trials. Blocks 1–7 only contained ‘non-switch’ trials where the proportion of elements remained constant throughout the trial. The first block contained 20 practice trials where 75% of the elements in the grid were the same color (in ten trials 75% of the elements were pink and in ten trials 75% of the elements were blue). Participants received feedback on their responses in this block. This feedback was displayed for 500 ms, and consisted of “correct”, “wrong”, or “invalid response” for invalid key presses. Feedback was followed by a blank screen for 250 ms. In blocks 2–22, participants did not receive feedback. The second block contained 72 additional practice trials where 54% of the elements in the grid were the same color. In the third block, participants completed 72 trials, which were composed of 24 trials at three different difficulty levels: 52, 54, or 56% of the elements had the same color. At the conclusion of the third block, one of the three difficulty levels was selected for the remainder of the experiment using the following algorithm to account for individual differences in ability so that performance was away from floor and ceiling:
-
1)
If the participant’s accuracy was exactly 75% for a specific difficulty level, then this level was selected. If more than one difficulty level achieved 75% accuracy, then the hardest level was selected.
-
2)
If no difficulty level achieved accuracy of exactly 75%, then the level with accuracy higher and closest to 75% was selected. If there was a tie between difficulty levels, then the hardest one was selected.
-
3)
If none of the difficulty levels achieved 75% accuracy, then the easiest level (i.e., 56%) was selected.
Blocks 4–7 each contained 72 trials with the difficulty level selected at the end of block 3. These four blocks provided the participant with additional practice at the selected difficulty level and helped control for learning effects before the main part of the experiment. The response times in block 7 were used to determine the ‘switch time’ for the remainder of the experiment. On switch trials, the change in color proportion occurred at the switch time, which was set to the median RT of block 7 minus 250 ms to account for response-production time. Blocks 8–22 were the main experiment and consisted of half non-switch trials and half switch trials. The two trial types were randomly ordered.
Blocks 23 and 24 tested whether participants could detect changes in color proportions. During these blocks, they were instructed to only respond if the proportion of colored elements changed. They were told to withhold responses if the proportion remained constant. Block 23 contained 24 practice trials with feedback. At the end of each trial, participants saw a message “correct”, “wrong”, or “invalid response” displayed for 500 ms. Block 24 contained 72 trials with no feedback. In both blocks, half of the trials were switch trials and the other half were non-switch trials, randomly ordered. The switch time and difficulty level for these blocks was the same as in the main task.
Modeling results
The model we fit to this data set is a hierarchal extension of the pDDM model described previously. Given the simple structure of this experiment where all participants come from a single population, we assume that the hierarchal structure consists of a single group. Based on this, we make the standard assumption that individuals are members of a normally distributed population and assign a normal prior for each individual level parameter
where T N(μ, σ, a, b) refers to the normal distribution with mean μ and standard deviation σ truncated to the interval [a, b]. We further specify the following mildly informative priors for the hyper mean
and hyper standard deviation parameters respectively
Here, Γ(a, b) refers to the gamma distribution with shape and rate parameters a and b, respectively.
To fit this hierarchal model, we utilize the same PDA method embedded into the DEMCMC framework as discussed above with the same values for the methodological parameters (N s , h, d t). Figure 6 shows the posterior distributions for the hyper mean parameters μ a , μ z , μ 1, μ 2, μ N D , μ d e l a y . First, the rate of evidence accumulation after the change of information (μ 2) is the opposite sign of that before, as expected. Second, the delay parameter has a mean value of ∼ 440 ms, which is consistent with the large delay found in Holmes et al. (2016) using a non-stationary variant of the RDM task. Finally, we again note that the posterior distributions for μ 2 and μ d e l a y are not as well constrained as those for other parameters. As with the parameter recovery experiments above, this is a result of the relative sparsity of participant responses that occur after the stimulus change. Additionally, posteriors for μ s z and μ s μ (fit but not shown) were not well constrained.
To assess quality of fit, we plot the actual and predicted choice probability and mean RT for all participants in Fig. 7 as well as the full RT distribution for an indicative participant. Choice and mean RT plots show good agreement and the participant RT distribution fit (Fig. 7c) shows good agreement, particularly given the bimodality of the RT distribution for choice 2.
Quality of fit. a, b Comparison of model predictions for the mean choice probabilities and mean response times against data. The straight lines represent the y = x axis, which would correspond to perfect agreement. To perform this comparison, 10,000 independent stochastic simulations of the pDDM with and without a switch were performed using the mean values of the posterior parameter distribution. The fraction of simulations corresponding to the different choice alternatives and mean response time were recorded for each switch and non-switch trials separately. In these figures, “Choice 1” (C1) refers to the first stimulus presented while C2 refers to the second stimulus presented. In (a), the choice probabilities are reported for C1 while in (b), the mean RT is reported for both C1 and C2 responses on both switch and non-switch trials. c Comparison of RT distribution for the data (dot) and the model based prediction (line) derived from the mean of the posterior distribution for a representative individual
Technical notes
Analysis of likelihood approximation errors for DDM
While the PDA is a powerful method of performing Bayesian parameter estimation, it is important to keep in mind that the likelihood approximation used is just that, an approximation. As with any numerical method, it is important to assess the size of estimation errors and how they may influence results. Above, this method was used to fit a piecewise variant of the canonical DDM model with piecewise constant, time-varying drift rates. Here, we investigate how the methodological parameters (h, N s ) influence the accuracy of likelihood estimation for the simpler DDM model. Additionally, since this method requires the simulation of stochastic differential equations (SDE), we also assess the influence of the time discretization parameter dt on accuracy.
Each of these factors has been investigated in previous works. Numerous automated selection criteria for (h, N s ) for general KDE applications have been proposed in the past. However, to our knowledge, there is no widely accepted gold standard at this time. We thus perform an analysis of their influence in order to make an informed selection. Similarly, the method of simulating stochastic evidence accumulation models (such as the DDM) and the influence of time step size (d t) was investigated in Brown et al. (2006). As we will see, however, these tuning parameters are intertwined, and they must be chosen jointly rather than independently.
To evaluate the influence of these methodological “tuning” parameters, we use the standard DDM model discussed above. We begin by first generating a baseline data set of 1,000 observations from the DDM model with a fixed set of parameters (see Fig. 2 caption for those parameters, chosen in accord with values in Matzke and Wagenmakers (2009) and Ratcliff and McKoon (2008)). To generate this data set, we use a first-order Euler Maruyama (EM) scheme (Higham 2001) to simulate the drift diffusion equation
where y denotes the level of evidence or activation value associated with a decision and dw is a standard white noise with mean 0 and variance of 1. Letting y(t i ) denote the value of y on a grid of time points {t i } (where t i+1 = t i + d t), the EM scheme is simply the stochastic version of the well-established Euler scheme for simulating ordinary differential equations and involves iteratively updating y according to
where η is drawn from the standard normal distribution. To generate this data set, we use a very fine value of d t = 0.0001. The dots in Fig. 2 show the response time distribution for correct and incorrect responses for this simulated data set.
Next, we use this baseline data set to determine how (N s , h, d t) influence the accuracy of the likelihood approximation. To do so, we fix the parameters that generated this data set. For each value of (N s , h, d t), we generate N s choice/RT pairs from the model by simulating Eq. (15) with dt. Next, the KDE of this synthetic data set is computed using a Gaussian kernel (K) (as in Holmes, 2015) with the smoothing parameter h. To determine the influence of methodological parameters on accuracy, one needs to compare the approximate density function to the “exact” density function. Unfortunately for DDM, no such “exact” function exists. Solutions of the DDM first passage time problem have been worked out, however they either involve approximating infinite series (Navarro and Fuss 2009), numerical integrals (Tuerlinckx 2004), or solutions to partial differential equations (Voss and Voss 2008). In lieu of an exact likelihood function to compare against, we generate a much higher quality approximation with N s = 500,000, h = 0.0005, and d t = 0.0001. Figure 8 shows a comparison of the integrated squared error (ISE, which is a common measure of how close two functions are to each other) and the log likelihood (with respect to the 2000 baseline observations) evaluated with (N s , h, d t) and the much more intensive approximation respectively (which takes the place of the exact solution).
Error analysis of DDM using the PDA method. Errors in the PDA estimation of the DDM choice/RT probability density as a function of simulation sample number (N s ), smoothing bandwidth (h, denoted in s), and simulation time step (dt). Each panel represents a different combination of (N s , h) while the horizontal axis screens different values of d t = 0.001,0.002,0.004,0.008,0.016 s. For each set of (N s , h, d t), ten replicant simulations were performed and average errors are reported. Since we do not have access to the true likelihood function, measures of error are computed by comparing the approximation at each combination of methodological parameters to a separate, more accurate approximation of the true density function obtained with N s = 500,000 samples, a smoothing bandwidth of h = 0.001 s, and a time step of d t = 0.0001 s. The gray curves indicate the integrated squared error, obtained by comparing the approximate and more accurate distributions at 100 evenly spaced time points in the response time interval of 0.15–2 s. To determine the accuracy of log-likelihood estimates (indicated in black), a 5000 sample choice/RT data set (mimicking the data set of an experimental participant) was generated, log-likelihood values were computed using the presented combinations of (N s , h, d t), and those values were compared to a more accurate approximation. The horizontal scale is logarithmic and the vertical scale is linear on all plots. Model parameters are the same as in Fig. 2. All errors reported are percent errors
These results demonstrate that for all combinations of methodological parameters, log-likelihood estimation errors are on the order of 1 − 5% with ISE < 3%. Increasing the number of samples (N s ), decreasing the smoothing bandwidth (h), and decreasing the SDE time step size (d t) all of course lead to improved accuracy. However, in each case, there is a point of diminishing returns. Increasing N s or decreasing dt both lead to a linear increase in computation time; doubling either doubles the computation time. Additionally, decreasing h leads to increased variance in the likelihood estimation, which can degrade the performance of an MCMC algorithm (Holmes 2015). Thus, one must balance performance with accuracy.
These results also demonstrate the intertwined nature of these methodological parameters. For example, a very refined value of dt provides little benefit if the smoothing parameter h is large. Intuitively, this is sensible. A more refined stochastic simulation provides a better representation of the fine-grained features of the model. The KDE, however, essentially smoothes the model likelihood function. Thus, the extra refinement gained through a reduction in dt would be smoothed over in a sense if h is not commensurately small. Based on these results, it appears a choice of d t = 0.5 ⋅ h is reasonable. From here on, we choose values N s = 10,000, h = 0.02 sec, and d t = 0.01 sec. This yields an estimation error of < 2%.
Timing
A central feature of this PDA methodology is that in order to determine how well a particular parameter set accounts for observed data, a large number (N s ) of model simulations must be generated for comparison. Consider the case of the DDM model discussed above. At each parameter set, N s = 10,000 model simulations is performed. In order to estimate the posterior distribution, 18 MCMC chains (three times the number of parameters as is standard for DEMCMC) are run simultaneously for 2,500 MCMC iterations. The whole procedure thus requires 450 million simulations of the diffusion process described by Eq. (14). Thus computation time must be considered when using this method.
From a methodological perspective, there are a number of choices that must be made to balance performance and approximation accuracy. First, the level of temporal resolution in the SDE simulation (dt) linearly increases computational time. That is, halving the time step, which doubles the number of time steps required, doubles the simulation time. Similarly, computation time linearly increases with the number of simulation samples (N s ) used to approximate the density function; doubling N s doubles computation time. These factors must be considered to ensure that the models being generated can be fit within a reasonable timeframe.
Some efficiencies can be leveraged to improve performance of this method (which have been here and are implemented in the codes used to generate these timings). Built-in functions in common programming languages such as Matlab and R can be called to perform the kernel density estimation. However, as discussed in Holmes (2015), these can be inefficient, and custom routines can greatly improve their performance. Furthermore, while these approximate likelihood values can be plugged directly into a standard MH algorithm, doing so can lead to poor MCMC acceptance rates. This can, however, be ameliorated by a simple adjustment to the MH algorithm (likelihood resampling, detailed in Holmes (2015)).
On a standard desktop computer (2015 iMac) utilizing four compute cores in parallel, this fitting of the simple DDM model takes ∼ 40 minutes. Fitting the pDDM model necessitates an increase in the number of chains from 18 to 24 (since there are eight parameters to be fit versus six for DDM). Since computation time grows linearly with the number of chains, this yields a roughly 1/3 increase in computation time. In the case of the pDDM model described here, we have the added complexity of having both stationary (where the stimulus does not change) and non-stationary trials (where it does change). Since these represent different trial types, N s = 10,000 samples must be simulated to approximate a density function for each trial type, yielding a doubling of the computation time. This demonstrates that computation time will be determined by not only the model but also the experimental setup since each unique experimental condition requires a separate set of associated simulation conditions. In Holmes et al. (2016), for example, participants were shown 15 unique trial types throughout the course of the experiment, necessitating the generation of N s model simulations for each of those 15 trial types in order to fit the pLBA model using this method. Thus, computational limitations must be considered when initially designing experiments if this method is to be used.
The test case used to demonstrate this method involved fitting data derived from 15 participants, each of which performed a moderately large number of trials of two types: (1) stationary where the stimuli did not change over time and (2) non-stationary where the stimulus did change over time. Using N s = 10,000 simulations per trial condition, 24 chains, and 2500 MCMC iterations yields 18 billion individual SDE simulations needed to fit this data set. Fortunately, the recent simplification of parallel computer architectures provides easy avenues to speed this process. For this application, we utilize Matlab and its built-in parallel computing toolbox to distribute this computation to eight computing cores at the Advanced Computing Center for Research and Education (ACCRE, at Vanderbilt University) to accelerate this fit. Using this facility, the hierarchal fit of all 15 participants took approximately 2 days.
While these computation times may seem daunting, we note that the ever-improving performance of and access to parallel computing facilities will make the use of this methodology more feasible over time, provided the interested researcher takes care in designing models and experiments with these constraints in mind. For example, migrating this application from eight cores to larger more powerful machines (32-core desktops, which are readily available and affordable these days) can reduce implementation times by a factor of 3–4. Additionally, increasingly user-friendly access to general-purpose GPU computing resources hold the potential for yet more dramatic increases in performance, which over time will allow researchers to utilize this method and these models with fewer constraints on model/experimental design as well as improved accuracy.
General discussion
Here, we describe a model, the piecewise diffusion decision model, in combination with a Bayesian fitting methodology (the PDA) that can be used to model complex decision tasks with non-stationary inputs. Given the time-heterogeneous nature of this model, the lack of tractable analytic descriptions of the likelihood function, and the approximate nature of the PDA, we first set out to verify that the combination of the model and fitting methodology are sufficient to recover parameters of simulated data. Results of fitting the simpler DDM and pDDM models show this to be the case, with some caveats, which will be described momentarily.
With this methodology established, we fit the pDDM model to new experimental data to examine how a single change of information during the course of deliberation influences choices. Results show that there is a substantial delay, on the order of 400–500 ms, between the presentation of new information and an influence of that information on evidence accumulation. This is consistent with results from Holmes et al. (2016) where a similar 400–500 ms integration delay was found, suggesting that this model is an effective tool for measuring how changes of information influence decision processes. To be clear, we are not suggesting that a discrete and delayed neural change in accumulation occurs when information changes. Rather, there is likely a nonlinear process involving visual short-term memory or inhibition that leads to a delayed influence of new information.
While this current methodology is very promising, there are a couple of important caveats. The first caveat is that trial-to-trial variability (related to bias and drift rates) parameters are not well estimated in this approach. While trial-to-trial drift variability is known to be important for accounting for long errors in RT distributions, these parameters are rarely the primary source of inferences and thus are not the focus here. The second is that parameter recovery is not perfect. The peak of posterior distributions do not align perfectly with the parameter values generating the simulation data (though the exact values are well within the credible intervals). This is not a result specifically of the PDA method though and the imprecision in part results from the nature of Bayesian methods, noise in data, and the approximate nature of MCMC in general. Further, some parameters exhibit a larger spread in their posterior distributions due to the structure of the data available, indicating less confidence in their estimates. As discussed in Holmes (2015), this results from the approximate nature of the modeling process, the intrinsic “sloppiness” of models (Apgar et al. 2010; Gutenkunst et al. 2007), and arises in other fitting methodologies such as maximum likelihood estimation (Brown et al. 2006).
As has been noted in numerous other fields such as mathematical biology, most models containing more than a few parameters exhibit “sloppiness”, which describes models where a wide range of parameter values can nearly equally account for observed data. At its most basic level, sloppiness is the result of having data that is insufficient to directly constrain the parameters of a model. This insufficiency often comes from the inability to observe elements of a model that one wants to constrain. In a biological context, this is usually an issue when one tries to infer the properties of a signaling network with only data about the input and output of that network. With no observations of the complex network of biological molecules or intermediaries that translate an input into an output, it is nearly impossible to quantitatively constrain the properties of those intermediaries with input/output data alone.
Making inferences about a decision process raises similar issues. Where evidence accumulation models are concerned, a data set usually consists of information about the stimulus input (easy versus hard for example) and the response output (choice/response time). From the perspective of the model, this is a very sparse measurement since numerous aspects of the accumulation process are not observed (bias, threshold, accumulation rate, non-decision time) and are only indirectly constrained by the data. The issue of sloppiness is somewhat exacerbated when approximations to a model must be made (as is the case with pDDM). Statistically speaking, sloppy models are those for which a range of parameter values lead to a similar likelihood. In this case, the small estimation errors introduced by approximations, whether it be summary statistics in ABC methods, truncation of series that are often involved in calculation of DDM likelihoods, SDE simulation of more complex models, or the KDE approximation used in the PDA, can skew the posterior toward one part of the sloppy region of parameter space.
We stress, however, that this is an issue of parameter recovery, not of model fit, as noted in Brown et al. (2006) and Holmes et al. (2016). Our results show that even when model parameters are not perfectly recovered or tightly constrained, the posterior mean parameters still provide excellent fits in both parameter recovery experiments. Thus, while one must take care when making direct, quantitative comparison of parameters between conditions or across experimental populations, this modeling platform allows the user to determine how well the assumptions encoded in the model account for experimental data.
While we used pDDM to understand how a single change of information influences decisions, what is presented here is a modeling platform rather than a single model. Since this is a simulation-based approach, it provides the researcher freedom to consider models containing elements not previously tractable due to the difficulty in deriving analytic approximations to likelihood functions. For example, this approach could be used to investigate theories involving collapsing bounds (Resulaj, Kiani, Wolpert, & Shadlen, 2009), urgency gating (Cisek, Puskas, & El-Murr, 2009; Thura, Beauregard-Racine, Fradet, & Cisek, 2012), or neural inhibition (Usher and McClelland 2001; Turner et al. 2014). Multiple changes of information could be incorporated as might be of interest if multiple choice alternatives are present that people sequentially attend to. Furthermore, it opens up the possibility of closely linking temporal observations (such as eye tracking) to models to investigate how the changing state of the decision-maker (what they are looking at when for example) influences decisions. Given the increasing power and ease of computational technologies, we hope this approach will expand the applications of sequential sampling models to more complex decision tasks.
Notes
There is currently a multi-lab collaboration underway led by Udo Boehm, University of Groningen, to assess this issue (personal communication).
References
Apgar, J. F., Witmer, D. K., White, F. M., & Tidor, B. (2010). Sloppy models, parameter uncertainty, and the role of experimental design. Molecular BioSystems, 6(10), 1890–1900.
Bröder, A., & Schiffer, S. (2006). Adaptive flexibility and maladaptive routines in selecting fast and frugal decision strategies. Journal of Experimental Psychology: Learning, Memory, and Cognition, 32(4), 904–918.
Brown, S. D., Ratcliff, R., & Smith, P. L. (2006). Evaluating methods for approximating stochastic differential equations. Journal of Mathematical Psychology, 50(4), 402–410.
Busemeyer, J. R., & Townsend, J. T. (1993). Decision field theory: A dynamic-cognitive approach to decision-making in an uncertain environment. Psychological Review, 100(3), 432–459.
Cisek, P., Puskas, G. A., & El-Murr, S. (2009). Decisions in changing conditions: The urgency-gating model. The Journal of Neuroscience, 29(37), 11560–11571.
Diederich, A. (1997). Dynamic stochastic models for decision-making under time constraints. Journal of Mathematical Psychology, 41(3), 260–274.
Diederich, A., & Busemeyer, J. R. (1999). Conflict and the stochastic-dominance principle of decision-making. Psychological Science, 10(4), 353–359.
Diederich, A., & Oswald, P. (2016). Multi-stage sequential sampling models with finite or infinite time horizon and variable boundaries. Journal of Mathematical Psychology, 74, 128–145.
Diederich, A., & Trueblood, J. (submitted). A dynamic dual process model of risky decision-making.
Gold, J. I., & Shadlen, M. N. (2001). Neural computations that underlie decisions about sensory stimuli. Trends in Cognitive Sciences, 5(1), 10–16.
Guo, L., Trueblood, J. S., & Diederich, A. (2015). A dual-process model of framing effects in risky choice. In Noelle, D.C., & et al (Eds.), Proceedings of the 37th annual conference of the cognitive science society (pp. 836–841). Austin, TX: Cognitive Science Society.
Gureckis, T. M., & Love, B. C. (2009). Learning in noise: Dynamic decision-making in a variable environment. Journal of Mathematical Psychology, 53(3), 180–193.
Gutenkunst, R. N., Waterfall, J. J., Casey, F. P., Brown, K. S., Myers, C. R., & Sethna, J. P. (2007). Universally sloppy parameter sensitivities in systems biology models. PLoS Computational Biology, 3(10), e189.
Higham, D. J. (2001). An algorithmic introduction to numerical simulation of stochastic differential equations. SIAM Review, 43(3), 525–546.
Holmes, W. R. (2015). A practical guide to the probability density approximation (PDA) with improved implementation and error characterization. Journal of Mathematical Psychology, 68, 13–24.
Holmes, W. R., Trueblood, J. S., & Heathcote, A. (2016). A new framework for modeling decisions about changing information: The piecewise linear ballistic accumulator model. Cognitive Psychology, 85, 1–29.
Krajbich, I., & Rangel, A. (2011). Multialternative drift-diffusion model predicts the relationship between visual fixations and choice in value-based decisions. Proceedings of the National Academy of Sciences, 108(33), 13852–13857.
Krajbich, I., Armel, C., & Rangel, A. (2010). Visual fixations and the computation and comparison of value in simple choice. Nature Neuroscience, 13(10), 1292–1298.
Logan, G. D., & Burkell, J. (1986). Dependence and independence in responding to double stimulation: A comparison of stop, change, and dual-task paradigms. Journal of Experimental Psychology: Human Perception and Performance, 12(4), 549–563.
Matzke, D., & Wagenmakers, E.-J. (2009). Psychological interpretation of the ex-Gaussian and shifted Wald parameters: A diffusion model analysis. Psychonomic Bulletin and Review, 16(5), 798–817.
Navarro, D. J., & Fuss, I. G. (2009). Fast and accurate calculations for first-passage times in Wiener diffusion models. Journal of Mathematical Psychology, 53(4), 222–230.
Ratcliff, R. (1978). A theory of memory retrieval. Psychological Review, 85, 59–108.
Ratcliff, R. (1980). A note on modeling accumulation of information when the rate of accumulation changes over time. Journal of Mathematical Psychology, 21(2), 178–184.
Ratcliff, R., & McKoon, G. (2008). The diffusion decision model: Theory and data for two-choice decision tasks. Neural Computation, 20(4), 873–922.
Ratcliff, R., Thapar, A., & McKoon, G. (2001). The effects of aging on reaction time in a signal detection task. Psychology and Aging, 16(2), 323–341.
Resulaj, A., Kiani, R., Wolpert, D. M., & Shadlen, M. N. (2009). Changes of mind in decision-making. Nature, 461(7261), 263–266.
Shadlen, M. N., & Newsome, W. T. (1996). Motion perception: Seeing and deciding. Proceedings of the National Academy of Sciences, 93(2), 628–633.
Silverman, B. W. (1982). Algorithm as 176: Kernel density estimation using the fast Fourier transform. Journal of the Royal Statistical Society. Series C (Applied Statistics), 31(1), 93–99.
Silverman, B. W. (1986). Density estimation for statistics and data analysis (Vol. 26). CRC Press.
Smith, P. L., & Ratcliff, R. (2004). Psychology and neurobiology of simple decisions. Trends in Neurosciences, 27(3), 161–168.
Srivastava, V., Feng, S. F., Cohen, J. D., Leonard, N. E., & Shenhav, A. (2017). A martingale analysis of first passage times of time-dependent Wiener diffusion models. Journal of Mathematical Psychology, 77, 94–110.
Ter Braak, C. J. (2006). A Markov chain Monte Carlo version of the genetic algorithm differential evolution: Easy Bayesian computing for real parameter spaces. Statistics and Computing, 16(3), 239–249.
Thura, D., Beauregard-Racine, J., Fradet, C.-W., & Cisek, P. (2012). Decision-making by urgency gating: Theory and experimental support. Journal of Neurophysiology, 108(11), 2912–2930.
Tsetsos, K., Gao, J., McClelland, J. L., & Usher, M. (2012). Using time-varying evidence to test models of decision dynamics: Bounded diffusion vs. the leaky competing accumulator model. Frontiers inNeuroscience, 6.
Tsetsos, K., Usher, M., & McClelland, J. L. (2011). Testing multi-alternative decision models with non-stationary evidence. Frontiers in neuroscience, 5.
Tuerlinckx, F. (2004). The efficient computation of the cumulative distribution and probability density functions in the diffusion model. Behavior Research Methods, Instruments, & Computers, 36(4), 702–716.
Turner, B. M., & Sederberg, P. B. (2014). A generalized, likelihood-free method for posterior estimation. Psychonomic Bulletin & Review, 21(2), 227–250.
Turner, B. M., Sederberg, P. B., Brown, S. D., & Steyvers, M. (2013). A method for efficiently sampling from distributions with correlated dimensions. Psychological Methods, 18(3), 368–384.
Turner, B. M., Sederberg, P. B., & McClelland, J. L. (2014). Bayesian analysis of simulation-based models. Journal of Mathematical Psychology, 72, 191–199.
Turner, B. M., Van Maanen, L., & Forstmann, B. U. (2015). Informing cognitive abstractions through neuroimaging: The neural drift diffusion model. Psychological Review, 122(2), 312–336.
Usher, M., & McClelland, J. L. (2001). The time course of perceptual choice: The leaky, competing accumulator model. Psychological Review, 108(3), 550–592.
Vandekerckhove, J., & Tuerlinckx, F. (2007). Fitting the Ratcliff diffusion model to experimental data. Psychonomic Bulletin & Review, 14(6), 1011–1026.
Vandekerckhove, J., & Tuerlinckx, F. (2008). Diffusion model analysis with MATLAB: A DMAT primer. Behavior Research Methods, 40(1), 61–72.
Vandekerckhove, J., Tuerlinckx, F., & Lee, M. D. (2011). Hierarchical diffusion models for two-choice response times. Psychological Methods, 16(1), 44–62.
Verbruggen, F., & Logan, G. D. (2008). Response inhibition in the stop-signal paradigm. Trends in Cognitive Sciences, 12(11), 418–424.
Voss, A., & Voss, J. (2007). Fast-dm: A free program for efficient diffusion model analysis. Behavior Research Methods, 39(4), 767–775.
Voss, A., & Voss, J. (2008). A fast numerical algorithm for the estimation of diffusion model parameters. Journal of Mathematical Psychology, 52(1), 1–9.
Wabersich, D., & Vandekerckhove, J. (2014). Extending jags: A tutorial on adding custom distributions to jags (with a diffusion model example). Behavior research methods, 46(1), 15–28.
White, C. N., Ratcliff, R., Vasey, M. W., & McKoon, G. (2010). Using diffusion models to understand clinical disorders. Journal of Mathematical Psychology, 54(1), 39–52.
Wiecki, T. V., Sofer, I., & Frank, M. J. (2013). HDDM: Hierarchical Bayesian estimation of the drift-diffusion model in Python. Frontiers in Neuroinformatics, 7, 14.
Acknowledgments
WRH and JST were supported by National Science Foundation Grant SES-1556325.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Holmes, W.R., Trueblood, J.S. Bayesian analysis of the piecewise diffusion decision model. Behav Res 50, 730–743 (2018). https://doi.org/10.3758/s13428-017-0901-y
Published:
Issue Date:
DOI: https://doi.org/10.3758/s13428-017-0901-y