Abstract
Background
Genetic variation among different strains of wild macrofungi can be expressed and documented using different molecular tools. In this study, the genetic diversity and relatedness of Pleurotus ostreatus from different locations in Ondo and Ekiti States were investigated.
Methods
Random amplified polymorphic DNA-polymerase chain reaction marker was adopted to assess the genetic diversity of some wild P. ostreatus. Microorganisms associated with the wild P. ostreatus and their soil (substrate) were enumerated and identified using standard microbiological methods, while composition of soil around the mushrooms was evaluated using the conventional method.
Results
A total of 114 positive DNA bands were observed among wild P. ostreatus based on random amplified polymorphic DNA analysis (RAPD) with 10 primers, showing a total number of polymorphic markers of 91 and average polymorphism of 80.24%. P. ostreatus from Ala quarters and Igbatoro Road in Akure related to P. ostreatus from Ado-Ekiti, while P. ostreatus from Ido-Ekiti and Usi-Ekiti are genetically similar. The bacterial count from wild P. ostreatus and associated soil ranged from 1.20 × 104 to 4.70 × 105 and 1.70 × 107 to 6.10 × 108 CFU g−1, respectively. The highest fungal count of 3.40 × 104 and 7.40 × 107 SFU g−1 were obtained for wild P. ostreatus and associated soil at Usi-Ekiti, which possesses the highest organic content (5.90%). Isolated microorganisms were Pseudomonas putida, Streptomyces spp., Trichoderma spp., Penicillium italicum and others. The highest crude fiber (25.04%) and protein content (24.07%) were obtained in wild P. ostreatus from Usi-Ekiti.
Conclusion
RAPD revealed the genetic relatedness and genetic diversity among studied wild P. ostreatus, indigenous to two sates in Southwestern Nigeria. This will improve the strain selection for further utilization and documentation.

Similar content being viewed by others
Background
Pleurotus species are predominant in both temperate and tropical parts of the world; it naturally grows on decaying woody materials as well as above soil [1]. They are devoid of chlorophyllous tissues and, hence, considered as primary decomposer of organic matter. Mushrooms play important ecological niche by decomposing complex organic matter when their mycelial network extend into the soil to absorb nutrients, thereby contributing to the improvement of soil composition [2, 3].
Generally, species of edible and medicinal mushrooms are gaining much attention for advance biotechnological uses, especially in pharmaceutical and agro allied companies. The biodiversity of Pleurotus spp. has created potential utilization with profound biological and economical values [4], but their genetic variability and taxonomic controversies still express some limiting factors [5]. However, exact characterization and identification of Pleurotus species is fundamental to reveal their true identity for maximum exploitation in food industries and for research documentation. The improvement in the use of deoxyribonucleic acid (DNA) for molecular technology has been proven to be a successful tool to solve taxonomic chaos [6]. Random amplified polymorphic DNA (RAPD) is one of the cheapest and quickest methods for assessing the variability at DNA level, being explicitly useful in intraspecific analysis and displaying the accurate genotypic identity of fungi [7].
Moreover, microbial ecology and substrate composition are other paramount concerns that affect the relatedness and nutrient contents of wild edible mushrooms. Soil microbial communities are extremely diverse; relatively to their organic contents, water activity and soil nutrient, physical and chemical properties [8]. The microbial succession on wild edible mushroom as well as their substrates is crucial for mushroom farmers to enhance productivity, eliminate post-harvest contamination and maintain quality product. Therefore, microbial community associated with wild mushrooms and their substrate (soil) has not been adequately investigated and revealed. Our study, thus, provides information on the molecular variability of some P. ostreatus from different locations and reveals the microorganisms associated with P. ostreatus as well as composition of soil around them.
Materials
Collection of wild Pleurotus species and soil around them
Wild edible mushrooms, Pleurotus spp., were picked from the forest in Ala quarters (116.8 g), Igbatoro Road (97.5 g) in Ondo State, Ido-Ekiti (124.2 g), Usi-Ekiti (145.8 g) and Ado-Ekiti (106.6 g) in Ekiti State, Nigeria (Fig. 1). The soil associated with the growth of wild P. ostreatus at different locations was collected using hand soil auger from the surface to a depth of 0–20 cm into sterile cellophane bags. The samples were transferred to the laboratory immediately for analysis.
Methods
Extraction of DNA using cetyl trimethyl ammonium bromide (CTAB)
Each of the mushroom samples (5 g) was air dried and ground in mill machine. The prepared mushroom sample (0.5 g) was transferred into Eppendorf tube; DNA extraction buffer (500 µl) consisting of 2% w/v of CTAB diluted in 100 mM Tris–HCl, 20 mM EDTA, 1.4 M of NaCl and β-mercaptoethanol (2% v/v) was added and incubated at 65 °C for 60 min [9]. Thereafter, the sample was allowed to cool and centrifuged at the speed of 4800g at 4 °C for 10 min. 0.5 ml of chloroform and isoamyl alcohol (24:1) were added and vortexed for 10 s, followed by spinning at 5400g for 10 min. The supernatant was transferred into a new Eppendorf tube and 0.5 ml of cold isopropanol was added to precipitate the DNA. The sample was kept in the freezer for 1 h and later spun at 5400g for 10 min. The supernatant was discarded and the pellet was washed with 70% v/v ethanol. The DNA was dried at room temperature (26 ± 1 °C) and resuspended in 100 μl of sterile distilled water. Thereafter, the DNA concentration was measured using a spectrophotometer (Beckman Instruments Inc., Fullerto CA, USA) at 260 nm.
Random amplified polymorphic DNA-polymerase chain reaction (RAPD-PCR) analysis
Ten (10) RAPD primers were used for the generation of polymorphic fragments of P. ostreatus strains (Table 1). Amplifications was performed on 25 μl reaction mixture consisting of the following: 2.5 μl of PCR buffer, 2.5 μl of 10 mM dNTP mix, 2.5 μl of MgCl2, 1.0 μl of DNA, 1.0 μl of 10 pM primer (Operon Technologies, USA), 1.0 μl of Taq polymerase (Boehringer, Germany) and 14.5 μl of distilled water. The reaction volume in Eppendorf tube was amplified in a thermal cycler (Applied Biosystems 9700). The cycling program for amplifying RAPD-PCR assays was: 1st cycle of 94 °C for 3 min for initial denaturation, followed by 45 cycles of 94 °C for 20 s for denaturation, annealing of primer at 37 °C for 20 s and extension at 72 °C for 40 s with a final extension at 72 °C for 7 min.
Gel electrophoresis of PCR amplicons
Agarose gel electrophoresis was used to determine the quality and integrity of DNA by size fractionation on agarose gel. Hence, amplification products were resolved by electrophoresis using 1.0% agarose gel in 100 ml, 0.5× TBE buffer: 20 mM Tris, 89 mM boric acid, 20 mM EDTA, pH 8.0. The gel was allowed to cool down, 10 µl of 5 mg ml−1 ethidium bromide was added and mixed together before pouring into an electrophoresis chamber set with the combs inserted. After the gel had solidified, 10 μl of each PCR amplification product was mixed with a 2 µl of 6× loading buffer in the well created on agarose gel. Electrophoresis was done at 100 V for 2 h and the integrity of the DNA was visualized, photographed under UV light and documented with a gel documentation system (UVP, USA).
Cluster analysis
The position of the RAPD bands was scored and transformed into a binary character matrix as “1” stands for the presence, while “0” stands for the absence of an RAPD band at a particular position. Using the NTSYS-pc version 2.02 K package [10], the transformed binary character matrix data are first transferred into the software data collection module, from which pairwise distance matrices are compiled. The genetic diversity parameters were determined using the output data and graphical module of the software, and a phylogenetic tree was created by the unweighted pair-group method arithmetic average (UPGMA) to group individuals into clusters [11].
Isolation of bacteria and fungi from wild P. ostreatus and soil
The stock of each sample was prepared by putting 2.0 g of freshly harvested mushroom and 5.0 g of soil into sterilized peptone water of 8 ml and 45 ml, respectively. 1.0 ml of each sample was serially diluted in different test tubes to obtain appropriate dilution. Isolation of bacteria and fungi was done using pour plate method [12]. An aliquot (1.0 ml) of each diluted sample was transferred into a Petri dish with addition of nutrient agar (Lab M Lancashire, UK) for bacteria and potato dextrose agar (Lab M, Lancashire, UK) for fungi. The plates were allowed to solidify, incubated at 37 °C for 24 h and 28 ± 2 °C for 48 h for bacteria and fungi, respectively. Thereafter, the discrete colonies formed on growth media were counted and recorded as colony forming units per gram (CFU g−1) for bacteria and spore-forming unit per gram (SFU g−1) for fungi. The microbial mixture was transferred to the edge of an agar plate with an inoculating loop and streaked out over the surface of fresh agar plate. Pure colonies obtained were maintained on agar slants at 4 °C.
Identification of isolated microorganisms from wild mushrooms and soil samples
The isolated bacteria from wild mushrooms and soil samples were stained and subjected to biochemical tests such as catalase, citrate utilization, methyl red, Voges Proskauer, nitrate reduction, sulfur reduction, indole production, motility (SIM), triple sugar iron (TSI) and other sugar fermentation according to Cappuccino and Sherman [13] and Cheesbrough [12]. The results of biochemical tests were interpreted and identification of bacteria was carried out according to Cowan and Steel [14]. Fungi spores were stained with drops of lactophenol and viewed under a microscope. The microscopic characteristics were interpreted according to Barnett et al. [15].
Determination of soil composition around the wild mushrooms
The total organic matter and total organic carbon were determined by Walkley and Black method through chromic acid digestion [16, 17] with slight modification. Briefly, 0.5 g of each air-dried sample was put into a conical flask. Ten milliliter (10 ml) of 0.167 potassium dichromate (K2Cr2O7) and 20 ml of concentrated sulfuric acid (H2SO4) were added to the soil. The content was stirred to ensure good mixing of the soil sample with the reagents. After 30 min in a fume cupboard, 200 ml of distilled water, 10 ml of concentrated H3PO4 and 1 ml of 0.16% diphenylamine (indicator) were added. The excess dichromate that was not reduced in the reaction was determined by volumetric titration with 1 N FeSO4 solution. The percentage of sand (0.06–2.0 mm), silt (0.002–0.06 mm) and clay (less than 0.002 mm) were read on a textural triangle to determine the soil texture.
The mineral contents of the soil and mushroom were determined using the method of AOAC [18]. Briefly, each sample of mushroom and soil (1.0 g) was digested with HNO3, H2SO4 and HClO4 (3:1:1, v:v:v) with addition of hydrogen peroxide (500 μl) at 200 °C. The resulting solutions were cooled with 10 ml of ultrapure water (Milli-Q system, Millipore, USA) and analyzed for magnesium (Mg), iron (Fe), calcium (Ca), and zinc (Zn) with atomic absorption spectrophotometer (Buck 201 VGP). Flame photometer (Jenway PFP 7, Staffordshire, UK) was used to determine the potassium (K) and sodium (Na) contents of the samples. Each of the mineral standard solutions (1000 μg/ml Merck, Germany) was used for the calibration of the atomic absorption spectrophotometer.
Proximate composition of wild mushrooms Pleurotus ostreatus
The proximate composition of dried mushrooms was determined according to the method of AOAC [18]. Briefly, the moisture content in the mushrooms was determined by air oven drying 5 g of each sample at 110 °C for 2 h. The ash content was determined by incinerating the mushrooms (5 g) at 550 °C for 4 h, while the crude fiber in the mushroom samples was determined by dilute acid and alkali hydrolysis. The fat content in Pleurotus mushrooms was determined by extracting with diethyl ether in a Soxhlet apparatus, and the crude protein in Pleurotus mushrooms was determined by using Kjeldahl method and multiplying by 4.38. The total carbohydrates in the mushroom samples were calculated by difference using the formula below:
Statistical analysis
Data obtained during the experiment were subjected to analysis of variance. Variables with significant effects were characterized using Duncan’s new multiple range test at p ≤ 0.05 with the aid of SPSS version 17.0, Chicago, Illinois, USA.
Results and discussion
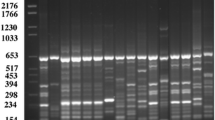
A total of 114 positive DNA bands were observed among the wild P. ostreatus based on RAPD analysis, showing total number of polymorphic markers of 91 and accounting for percentage of polymorphism ranging from 67 to 91% (Table 1). Typical RAPD gel photographs with marker bands of wild P. ostreatus are shown in Fig. 2. The outcome of Jacquard estimates with NTSYS using neighbor-joining cluster (Fig. 3) grouped the studied P. ostreatus into two clusters. Pleurotus ostreatus obtained from Ala quarters and Igbatoro Road in Akure labeled as 1 and 2, respectively, belonged to the same clade, showing 100% similarity. The harvested P. ostreatus from Ido-Ekiti (3) and Usi-Ekiti (4) are also genetically related. The pairing of P. ostreatus from Ado-Ekiti (5) in the same clade with P. ostreatus in Ala quarters and Igbatoro Road in Ondo State indicated a genetic relatedness. The RAPD cluster of Pleurotus species revealed both genetic relatedness and difference according to their geographical origins. The genetic relatedness could be attributed to the wide dispersal of fungi spore through a short- or long-distance distribution of Pleurotus species in the natural population across forest fragmentation, being promoted by human and animal movement [19].
Typical RAPD gel photographs with marker bands of wild P. ostreatus; a OPP 18, b OPS 15 and c OPQ-06, M: marker; 1: P. ostreatus from Ala quarters in Akure; 2: P. ostreatus from Igbatoro Road in Akure; 3: P. ostreatus from Ido-Ekiti; 4: P. ostreatus from Usi-Ekiti; and 5: P. ostreatus from Ado-Ekiti
Dendrogram of wild P. ostreatus from different geographic areas and labeled as 1: P. ostreatus from Ala quarters in Akure; 2: P. ostreatus from Igbatoro Road in Akure; 3: P. ostreatus from Ido-Ekiti; 4: P. ostreatus from Usi-Ekiti; and 5: P. ostreatus from Ado-Ekiti based on polymorphic RAPD markers showing genomic diversity of fungal strains using neighbor-joining cluster analysis method produced from Jacquard estimates with NTSYS Spc Version 2.02
The outcome of phylogenetic relation based on molecular analysis between different or similar populations of Pleurotus species provides a better resolution and understanding of their biogeography and speciation [5]. RAPD is a valuable method in distinguishing the different genotypes in mushrooms and evaluating their genetic similarities [7]. Other various biochemical and molecular techniques applied for genetic diversity, relatedness and identification of Pleurotus species were highlighted by Maftoun et al. [20] and Correa et al. [4]. These methods have been verified to be efficient in identification, expressed biodiversity in taxonomic system and unique in the selection of fungal strains.
The enumeration of bacteria (1.20 × 104 to 6.10 × 108 CFU g−1) and fungi (1.50 × 103 to 7.40 × 107 SFU g−1) from wild P. ostreatus and soil around P. ostreatus is presented in Table 2. The microbial count was more correlated to the organic matter than the genetic relationship exhibited by Pleurotus mushrooms. It was noted that soil from Usi-Ekiti possesses the highest microbial count: 7.40 × 107 SFU g−1 for fungi and 6.10 × 108 CFU g−1 for bacteria. The highest organic content (5.90%) in the location may require high microbial communities to improve nutrient cycle. Increase in soil bacteria and fungi creates microbial metabolic function to improve the soil biological characteristics [21]. The occurrence of isolated bacteria and fungi from P. ostreatus and their associated soil at different locations is shown in Tables 3 and 4. The presence of Pseudomonas putida conformed to the findings of Cho et al. [22] and Young et al. [23]. In their studies, they revealed the bacterium as one of the mushroom growth-promoting bacteria (MGPB) during the growth of P. ostreatus and Agaricus bisporus. Pseudomonas putida initiates primordial development and increases the yield of cultivated mushrooms [24], but Munsch and Alatossava [25] revealed that other members of the genus Pseudomonas exhibited saprophytic and pathogenic activities on A. bisporus and Pleurotus spp.
The species of Paecilomyces and Penicillium have been associated with white button mushroom in the study of Siyoum et al. [26]. The researchers revealed the microbial succession of compost, casing and mushroom using a plate count technique, denaturing gradient gel electrophoresis (DGGE) and sequencing of 16S and 18S rDNA. Trichoderma spp. was isolated with the highest occurrence from the examined P. ostreatus (33.30%) and their associated soil (17.10%). The cellulolytic filamentous fungi are responsible for green mold disease on mushrooms and spoilage of mushroom spawns [27]; the mold causes substantial and economic losses of some edible mushrooms, namely, species of Pleurotus, Agaricus and Lentinula in South Korea, Italy, Hungary, Canada, USA, Iran, New Zealand and Romania [28, 29]. Streptomyces spp., Bacillus subtilis, Actinomyces bovis, Pseudomonas putida, Penicillium italicum, Fusarium spp. and others were isolated from the soil around wild P. ostreatus. Streptomyces spp. produce indole-3-acetic acid (IAA), which acts as biocontrol against soil-borne pathogenic microorganisms. Many microorganisms exert growth-promoting effects by secreting beneficial secondary metabolites and enzymes that support spore germination and fruiting formation of wild mushrooms [30, 31]. The microbial ecology couples with optimal environmental conditions, and available nutrient or substrate composition are also selective parameters for the colonization and growth of mushrooms or other ectomycorrhizal fungi [32, 33].
In addition, soil microorganisms markedly influence soil composition by playing a key role in decomposing organic matter, cycling of carbon, nitrogen, phosphorous and stabilization of soil structure [34]. Therefore, the constituent of soil microorganisms could be a sensitive indicator of the growth of wild mushrooms.
The low occurrence and absence of some microorganisms from the examined mushrooms and soil may be due to shortcomings of traditional methods, that is, routine microbiological method adopted for microbial cultivation. This microbiological technique offers a narrow array for microbial isolation due to the fact that some microorganisms may remain viable but non-cultivable (VBNC). The method is only suitable for fast-growing and non-fastidious microbes, and overgrown fungi capable of producing large amounts of spores suppressing other fungi [35]. Another perception in relation to the low microbial occurrence could be associated with antibacterial, antifungal agents, lytic enzymes and volatile compounds secreted by Pleurotus mushrooms and their flora microorganisms, which contribute to the survival strategies of the edible fungi by reducing the occurrence of spoilage microorganisms, since mushrooms are often confronted by different types of bacteria, fungi and viruses [36, 37]. Hence, in associated ecosystem processes, a succession of microbial communities on wild mushrooms and their habitat (soil) involved intra- or interspecific interaction of different associations, which may include antibiosis, competition and mutualism [38].
Soil composition around wild Pleurotus mushrooms is presented in Table 5, while the nutrient contents of harvested wild P. ostreatus from different geographical locations in Ondo and Ekiti States are presented in Table 6. The sand content in the soil around P. ostreatus ranged from 66.30 to 88.33%. The clay contents of 10.30, 12.33 and 17.70% were obtained for soil collected around wild P. ostreatus at Ala quarters, Ado-Ekiti and Ido-Ekiti, respectively. They were higher (p ≤ 0.05) than those of Igbatoro (3.67%) and Usi-Ekiti (7.00%). The highest organic content (5.90%) in the soil around P. ostreatus mushroom harvested from Usi-Ekiti could contribute to its highest protein (24.07%) and crude fiber contents (25.04%). Nakalembe et al. [39] reported a high concentration of nutrient in edible mushrooms from the humid zone, which could be as a result of higher organic matter in the soil. Organic substances in the soil support high microbial biomass, metabolic activities and, thus, the assembly of more active nutrients into the food crops [40, 41]. The moisture, ash, dietary fiber, protein and carbohydrate contents in the harvested P. ostreatus mushrooms ranged from 7.30 to 10.19%, 6.26 to 8.45%, 19.05 to 25.04%, 0.85 to 1.77%, 17.30 to 24.07% and 30.06 to 45.10%, respectively. These values are within what were reported in the literature for proximate composition of Pleurotus spp. by the findings of Maftoun et al. [20]. Essential minerals, potassium (K), magnesium (Mg), calcium (Ca), iron (Fe) and zinc (Zn) in the studied mushrooms ranged from 71.08 to 221.06 mg/100 g, 27.00 to 111.40 mg/100 g, 40.90 to 101.78 mg/100 g, 26.10 to 81.03 mg/100 g and 10.71 to 43.00 mg/100 g respectively. The mineral contents of edible fungi may vary from one species to another based on their varieties, absorption mechanisms from the ecosystem, soil nutrient, mineral concentration in the substrates and environmental conditions [42]. Low fat (0.85–1.77%) and sodium (0.93–4.05 mg/100 g) contents were observed in the Pleurotus mushrooms. Several studies have indicated edible mushrooms as a better source of protein, dietary fibers, carbohydrate and essential minerals due to their ability to bioaccumulate nutrients into their fruit body with low or no cholesterol [43].
Conclusion
The study examined Pleurotus spp. indigenous to two states in Southwestern Nigeria and revealed their genetic relatedness and diversity. The use of molecular technology needs to be adopted to ascertain proper identification and documentation. The varying degrees in the microbial and nutrient contents of the studied wild P. ostreatus could be as a result of environmental factors, substrate constituent and geographical locations.
References
Chang ST, Miles PG. Mushrooms-cultivation, nutritional value, medicinal effect and environmental impact. 2nd ed. Boca Raton: CRC Press; 2004.
Cunningham AB, Yang X. Mushrooms in forests and woodlands: resource management, values and local livelihoods. London: Earthscan Ltd; 2011.
de Mattos-Shipley KMJ, Ford KL, Alberti F, Banks AM, Bailey AM, Foster GD. The good, the bad and the tasty: the many roles of mushrooms. Stud Mycol. 2016;85:125–57.
Correa RCG, Brugnari T, Bracht A, Peralta MR, Ferreira ICFR. Biotechnological, nutritional and therapeutic uses of Pleurotus spp. (Oyster mushroom) related with its chemical composition: a review on the past decade findings. Trends Food Sci Technol. 2016;50:103–17.
Menolli N, Breternitz BS, Capelari M. The genus Pleurotus in Brazil: a molecular and taxonomic overview. Mycosci. 2014;55:378–89.
Glen M. The use of DNA techniques to identify fungi. In: Potter K, Rimbawanto A, Beadle C, editors. Heart rot and root rot in tropical Acacia plantations. In: Proceedings of a workshop held in Yogyakarta, Indonesia, 7–9 February 2006. Canberra: ACIAR Proceedings;2006;124:46–54.
Park Y-J, Kwon O-C, Son E-S, Yoon D-E, Han W, Nam J-Y, Yoo Y-B, Lee C-S. Genetic diversity analysis of Ganoderma species and development of a specific marker for identification of medicinal mushroom Ganoderma lucidum. Afri J Microbiol Res. 2012;6(25):5417–25.
Cong J, Yang Y, Liu X, Lu H, Liu X, Zhou J, et al. Analyses of soil microbial community compositions and functional genes reveal potential consequences of natural forest succession. Sci Rep. 2015;5:10007–17.
Zolan ME, Pukkila PJ. Inheritance of DNA methylation in Coprinus cinereus. Mol Cell Biol. 1986;6:195–200.
Rohlf FJ. NTSYS-pc numerical taxonomy and multivariate analysis system, version 2.02. New York: Exeter Publication Setauket; 1998.
Sneath PHA, Sokal RR. Numerical taxonomy: the principle and practice of numerical classification. San Francisco: Freeman; 1973.
Cheesbrough M. District laboratory practice in Tropical Countries, part 2. Cambridge: Cambridge University Press; 2000. p. 12–69.
Cappuccino GJ, Sherman N. Microbiology, A laboratory manual; Biochemical activities of microorganism. 5th ed. California: Benjamin/Cumming science publishing; 1999.
Cowan ST, Steel KJ. Manual for the identification of medical bacteria. 3rd ed. Cambridge: Cambridge University Press; 1993.
Barnett JA, Payne RW, Yarrow O. Yeast characteristics and identification. 3rd ed. London: Cambridge University Press; 2000.
Sato JH, de Figueiredo CC, Marchão RL, Madari BE, Benedito LEC, Busato JG, de Souza DM. Methods of soil organic carbon determination in Brazilian savannah soils. Sci Agricol. 2014;71(4):302–8.
Walkley A, Black IA. An examination of Degtjareff method for determining soil organic matter and a proposed modification of the chromic acid filtration. Soil Sci. 1934;37:29–38.
Association of Official Analytical Chemists A.O.A.C. Official Methods of Analysis. Association of Official Analytical Chemists. 19th Gaithersburg, MD, USA; 2012.
Otieno OD, Onyango C, Onguso JM, Matasyoh LG, Bramwel W, Wamalwa M, Harvey JJW. Genetic diversity of Kenyan native oyster mushroom (Pleurotus). Mycolgia. 2015;107(1):32–8.
Maftoun P, Johari H, Soltani M, Malik R, Othman NZ, El Enshasy HA. The edible mushroom Pleurotus spp.: I. biodiversity and nutritional values. Int J Biotechnol Wellness Ind. 2015;4:67–83.
Yang Q, Wang X, Shen Y. Comparison of soil microbial community catabolic diversity between rhizosphere and bulk soil induced by tillage or residue retention. J Soil Sci Plant Nutri. 2013;13(1):187–99.
Cho Y-S, Kim J-S, Crowley DE, Cho BG. Growth promotion of the edible fungus Pleurotus ostreatus by fluorescent pseudomonads. FEMS Microbiol Lett. 2003;218:271–6.
Young L-S, Chu J-N, Hameed A, Young C-C. Cultivable mushroom growth-promoting bacteria and their impact on Agaricus blazei productivity. Pesq Agropec Bras. 2013;48(6):636–44.
Zarenejad F, Yakhchali B, Rasooli I. Evaluation of indigenous potent mushroom growth promoting bacteria (MGPB) on Agaricus bisporus production. World J Microbiol Biotechnol. 2012;28:99–104.
Munsch P, Alatossava T. Several pseudomonads, associated with the cultivated mushrooms Agaricus bisporus or Pleurotus sp. are hemolytic. Microbiol Res. 2002;157:311–5.
Siyoum NA, Surridge K, van der Linde EJ, Korsten L. Microbial succession in white button mushroom production systems from compost and casing to a marketable packed product. Ann Microbiol. 2016;66:151–64.
Kredics L, Jimenez LG, Naeimi S, Czifra D, Urbán P, Manczinger L, Vágvölgyi C, Hatvani L. A challenge to mushroom growers: the green mould disease of cultivated champignons. In: Mendez-Vilas A, editor. Current research, technology and education topics in applied microbiology and microbial biotechnology. Badajoz: Formatex Research Center; 2010. p. 295–305.
Colavolpe MB, Mejía SJ, Alberto E. Efficiency of treatments for controlling Trichoderma spp. during spawning in cultivation of lignicolous mushrooms. Bra J Microbiol. 2014;45(4):1263–70.
Hatvani L, Antal Z, Manczinger L, Szekeres A, Druzhinina IS, Kubicek CP, Nagy A, Nagy E, Vágvölgyi C, Kredics L. Green mold diseases of Agaricus and Pleurotus spp. are caused by related but phylogenetically different Trichoderma species. Phytopathol. 2007;97:532–7.
Kim MK, Math RK, Cho KM, Shin KJ, Kim JO, Ryu JS, Lee YH, Yun HD. Effect of Pseudomonas sp. P7014 on the growth of edible mushroom Pleurotus eryngii in bottle culture for commercial production. Bioresour Technol. 2008;99(8):3306–8.
Frey-Klett P, Burlinson P, Deveau A, Barret M, Tarkka M, Sarniguet A. Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Molecul Biol Rev. 2011;75(5):583–609.
Wang P, Liu Y, Yin Y, Jin H, Wang S, et al. Diversity of microorganisms isolated from the soil sample surround Chroogomphus rutilus in the Beijing region. Intern J Bio Sci. 2011;7(2):209–20.
Yamada A, Ogura T, Ohmasa M. Cultivation of mushrooms of edible ectomycorrhizal fungi associated with Pinus densiflora by in vitro mycorrhizal synthesis. Myco. 2001;11(2):59–66.
Loranger-Merciris G, Barthes L, Gastine A, Leadley P. Rapid effects of plant species diversity and identity on soil microbial communities in experimental grassland ecosystems. Soil Biol Biochem. 2006;38:2336–43.
Agrawal PK, Agrawal S, Shrivastava R. Modern molecular approaches for analyzing microbial diversity from mushroom compost ecosystem. 3 Biotech. 2015;5:853–66.
Lim Y, Ryu JS, Shi S, Noh W, Kim E, Le QV, Lee H-S, Ro H-S. Isolation of bacteria associated with the King Oyster mushroom, Pleurotus eryngii. Mycobiology. 2008;36(1):13–8.
Kim S-W, Kim S, Lee H-J, Park J-W, Ro H-S. Isolation of fungal pathogens to an edible mushroom, Pleurotus eryngii, and development of specific ITS primers. Mycobiology. 2013;41(4):252–5.
Boon E, Meehan CJ, Whidden C, Wong DH-J, Langille MGI, Beiko RG. Interactions in the microbiome: communities of organisms and communities of genes. FEMS Microbiol Rev. 2014;38:90–118.
Nakalembe I, Kabasa JD, Olila D. Comparative nutrient composition of selected wild edible mushrooms from two agro-ecological zones, Uganda. SpringerPlus. 2015;4:433.
Ortíz-Castro R, Contreras-Cornejo HA, Macías-Rodríguez L, López-Bucio J. The role of microbial signals in plant growth and development. Plant Signal Behav. 2009;4(8):701–12.
Vieira RF, Pazianotto RAA. Microbial activities in soil cultivated with corn and amended with sewage sludge. SpringerPlus. 2016;5:1844.
Zhu F, Qu L, Fan W, Qiao M, Hao H, Wang X. Assessment of heavy metals in some wild edible mushrooms collected from Yunnan Province, China. Environ Monit Assess. 2011;179(1–4):191–9.
Kalac P. Chemical composition and nutritional value of European species of wild growing mushrooms. In: Andres S, Baumann N, editors. Mushrooms: types, properties and nutrition. New York: Nova Science Publishers; 2012. p. 30–51.
Authors’ contributions
All the authors made substantial contributions to the conception, design and acquisition of the research study. TVF carried out the data collection and OCO did the interpretation. The manuscript was drafted by OCO and revised by all authors to meet up with the intellectual content. All authors read and approved the final manuscript.
Acknowledgements
The authors appreciate the technical support received from the Department of Microbiology, The Federal University of Technology, Akure, Nigeria.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All the data and materials are available for use.
Consent for publication
All the authors agreed to publish in the journal.
Ethics approval and consent to participate
This manuscript is an original research and has not been published in other journals.
Funding
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Familoni, T.V., Ogidi, C.O., Akinyele, B.J. et al. Genetic diversity, microbiological study and composition of soil associated with wild Pleurotus ostreatus from different locations in Ondo and Ekiti States, Nigeria. Chem. Biol. Technol. Agric. 5, 7 (2018). https://doi.org/10.1186/s40538-018-0119-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40538-018-0119-y