Abstract
The obligate intracellular Lawsonia intracellularis (LI), the etiological agent of proliferative enteropathy (PE), is an economically important disease in the swine industry. Due to extreme difficulty of in vitro culture of the pathogen, molecular characterization of protein components of LI that are targets of the immune system, is difficult; thus, the scientific evidence to drive the development of preventive measures is lacking. In this work, we investigated the antigenic and functional characteristics of a putative flagellar-associated protein, LI0570, using in silico computational approaches for epitope prediction and an in vitro protein-based molecular assay. The amino acid sequence of LI0570 exhibited similarities to flagellar-associated proteins in four different bacterial strains. The presence of B cell linear confirmative epitopes of the protein predicted by a bioinformatics tool was validated by western blot analysis using anti-LI mouse hyperimmune serum, which implied that LI0570 induced production of antigen-specific antibodies in vivo. Further, TLR5-stimulating activity and IL-8 cytokine expression produced via downstream signaling were observed in HEK-Blue™-hTLR5 cells stimulated with LI0570. This result indicates that the LI0570 protein can trigger an innate immune response followed by a T-cell-related adaptive immune response in an infected host. Collectively, the data presented here support that the LI0570 protein which shows the antigenic potential could be a useful component of a recombinant vaccine against PE, providing progress toward an effective prevention strategy.
Similar content being viewed by others
Introduction
Porcine proliferative enteropathy (PE) is caused by Lawsonia intracellularis (LI), an obligate intracellular bacterium, commonly occurring in swine herds. The infection results in thickening of the intestinal epithelium due to enterocyte proliferation. Clinical features such as chronic enteritis, lethargy, retarded growth rate, diarrhea, and acute hemorrhagic enteritis causing sudden death are exhibited. Indeed, Porcine PE is responsible for severe economic loss in the swine industry worldwide [1]. Proliferative enteropathy (PE) has also been diagnosed in a variety of animals such as horses, rabbits, rats, guinea pigs, dogs, chickens, sheep, deer, and non-human primates [2]. Despite high herd prevalence of LI infection in growing-finishing pigs, its pathogenic mechanisms remain speculative due to difficulty of in vitro culture of this obligate intracellular bacterium.
Intracellular motility of LI ultimately leads to cell dissemination, which enhances LI’s ability to penetrate mucous layers and colonize the intestinal tract. Other enteroinvasive bacteria pathogens such as Listeria and Shigella are randomly detected in the intestinal cell cytoplasm [3], while LI is largely present in the apical region of enterocytes [1]. Thus, molecular mechanisms of actin-based motility by which other intracellular bacterial pathogen spread in the infected cells may differ from those adapted by LI [4]. Interestingly, some enteric bacterial pathogens have mechanisms to penetrate the mucus layer to reach epithelial cells via flagella-driven motility which plays a role in the initial phase of infection [5, 6]. The presence of a single flagellar motor on LI, which is one of the main phenotypic elements of the pathogen, was observed in the supernatant of an infected cell culture in vitro [7] and also a preliminary analysis of the LI genome sequence (PHE/MN1-00; NCBI accession #NC_008011) exhibits genes responsible for flagellar assembly. It is also known that the bacterial protein flagellin effectively induces an innate immune response of the host that is mediated by its ability to bind to toll-like receptor 5 (TLR5). Flagellin, acting via TLR5, leads to activation of MyD88-dependent signaling and the proinflammatory transcription factor, NF-κB, which induces an intense innate and adaptive immune response against flagellated bacteria [8]. Although other bacterial flagellin has been widely used as an adjuvant molecule and as an antigen in vaccinology [9], the ability of LI flagellin to trigger the activation of immuno-modulatory pathway has not been demonstrated.
In this work, we initiated the study on the functional characteristics of LI0570 annotated by the United States National Center for Biotechnology Information (NCBI) as a putative flagellin and related hook-associated protein and its antigenic traits. We putatively defined LI0570 as Lawsonia flagellin (LFliC). Antigenic characteristics of LFliC were assessed by bioinformatics tools for in silico B-cell prediction. Further, we investigated whether the LI0570 retained immuno-adjuvant characteristics. To elucidate the role of the TLR5 agonist, TLR5-stimulating activity and IL-8 expression by the purified flagellin protein were measured using HEK293 cells.
Materials and methods
Bioinformatics analysis
Sequence analysis of the Lawsonia flagellin protein LI0570 was performed using the NCBI BLAST programs [10] and FASTA program [11]. Sequence similarity analysis was assessed by creating multiple sequence alignments using CLUSTALW [12]. The antigenicity index (linear B-cell epitopes) for the LFliC protein was estimated in silico using the BepiPred 2.0 web server [13]. BepiPred 2.0 employs the hidden Markov model combined with amino acid propensity scales to predict epitope data derived from crystal structures by assessing surface accessibility, helix probability, sheet probability, and coil probability [14].
Construction of the protein expression system
The predicted nucleotide sequences corresponding to amino acid sequences of LFliC (LI0570) were chemically synthesized (Bioneer, Daejeon, Korea). The synthesized genes were digested with EcoRI and HindIII endonucleases and subcloned into the Escherichia coli expression vector pET28a (+), which introduced a 6×His tag at the N-terminus of expressed proteins. The constructed plasmids pET28a-LfliC were transformed into E. coli BL21 (DE3) pLysS cells by electroporation and designated JOL1682. The His-tagged recombinant protein was purified from culture supernatant of JOL1682 strain expressing LfliC according to an affinity purification procedure using a nickel-charged nitrilotriacetic acid (Ni–NTA) resin as previously described [15]. Subsequently, the protein was quantified by Bradford assay. The bacterial strains and plasmids applied in this study are listed in Table 1.
Immunoblotting
Western blot analysis was performed to evaluate immunoreactivity of the LFliC protein in vitro using anti-LI hyperimmune serum obtained from the mice immunized with an L. intracellularis modified-live vaccine (Enterisol® Ileitis, Boehringer Ingelheim, Ingelheim am Rhein, Germany). The mice were inoculated with 5 × 106.9 50% tissue culture infectious doses (TCID50) via an oral route twice at 2-week intervals. The purified protein (10 μg/μL) was subjected to 15% SDS-PAGE and then transferred onto a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked with phosphate-buffered saline (PBS) buffer containing 3% bovine serum albumin (BSA) for 1 h, followed by incubation with the anti-LI mouse hyperimmune serum at a 1:300 dilution for 2 h. The antigen–antibody interaction was detected with a 1:5000 diluted horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Southern Biotechnology, Birmingham, AL, USA; diluted 1:5000).
In vitro characterization of LFliC protein
Determination of TLR5 bioactivity with the putative Lawsonia flagellin was conducted using HEK-Blue™-hTLR5 cells (InvivoGen, San Diego, CA, USA). The cells were incubated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), zeocin (50 μg/mL), and blasticidin (1 μg/mL). Cells (1 × 106 cells per well) that had been incubated overnight in a 96-well plates were stimulated with the LFliC protein (10 μg) in duplicate for various time periods (0, 1, 3, 4, and 7 h). The expression of TLR5 on the stimulated cells was assessed by fluorescence-activated cell sorting (FACS) analysis using an anti-TLR5 monoclonal antibody (1 μg/mL; Anti-hTLR5-IgG2a, Thermo Fisher Scientific, Waltham, MA, USA) as previously described [16]. Furthermore, expression of IL-8 cytokine produced by signaling through TLR5 was measured in the stimulated cells by reverse transcription-polymerase chain reaction (RT-PCR). The primers for the IL-8 gene were obtained previously [17]. Briefly, 1 × 106 HEK cells were treated with two concentrations of LFliC (10 and 100 ng/mL) in duplicate. The treated cells were incubated at 37 °C in a 5% CO2 incubator for 24 h and the transcription level of IL-8 was determined by RT-PCR as described previously [18].
In vitro cytotoxicity assay
The cytotoxic potential of the LFliC protein was assessed by cell viability assay with thiazolyl blue tetrazolium bromide (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; MTT) [19]. Cells from a rat intestinal epithelial cell line (IEC-18) were plated at a concentration of 1 × 106 and were stimulated with various concentrations of LFliC (0.1, 1, 5, and 10 ng/mL) in duplicate. The pulsed cells were incubated at 37 °C in a 5% CO2 incubator for 24 h, and then the cells were incubated for an additional 4 h with 5 mg/mL MTT. The bromide salt of MTT was absorbed by the cells and was reduced to a product called formazan within the mitochondria, which accumulated in the cell. The resulting blue formazan crystals was solubilized by dimethyl sulfoxide (DMSO), and the absorbance value was measured at 560 nm using a microplate spectrophotometer.
Statistical analysis
Non-parametric Mann–Whitney test was used to evaluate the statistical difference using GraphPad Prism (GraphPad Software Inc., La Jolla, CA, USA). P-values less than 0.05 were considered statistically significant.
Results
Characterization of LFliC as a putative antigen
Conserved domains of the LFliC (LI0570) protein in the L. intracellularis strain (PHE/MN1-00) were analyzed by searching NCBI’s Conserved Domain Database (CDD). The analysis showed that the protein contained conserved regions of flagellin and a related hook-associated protein FlgL (accession ID: COG1344; E-value, 4.38e−49) between amino acids 16–292 of the protein, a bacterial flagellin N-terminal helical region (accession ID: pfam00669; E-value, 2.40e−23) between amino acids 16–141 and a flagellar hook-associated protein (accession ID: TIGR02550; E-value, 7.74e−16) between amino acids 16–273. BLASTP similarity searches also revealed that the full amino acid sequence of LFliC (LI0570) of L. intracellularis strain (PHE/MN1-00) showed substantial similarity to the flagellin protein of Desulfovibrio alaskensis (Sequence ID: WP_011367650.1), Clostridium difficile (Sequence ID: YP_001086707.1), Pseudomonas aeruginosa (Sequence ID: NP_249783.1), and E. coli K-12 (Sequence ID: NP_416433.1) (Table 2). Sequence features of the multiple sequence alignment of LFliC to the flagellin-associated proteins of the four individual bacterial strains are shown in Figure 1. The identity varied among the strains at the protein levels (Table 2), with regions of extensive similarity between amino acids 1–151 and 202–294 of the LFliC sequence (Figure 1). This in silico characterization and analysis indicated that a putative Lawsonia flagellin protein, LI0570, shares a large degree of full-length amino acid sequence homology (40–77% identical residues) to the defined proteins as flagellin in other bacterial organisms (Table 2).
Sequence alignments of LI0570 and other flagellin-associated proteins using the CLUSTALW alignment program. An asterisk (*) indicates the amino acid residues that are identical in all sequences in the alignment, a colon (:) indicates conserved substitutions, and a period (.) indicates semi-conserved substitutions.
In vitro and in vivo characterization of immunogenic proteins
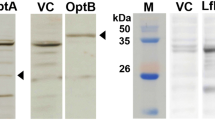
To evaluate potential antigenic features of the LI0570 protein, potential linear B-cell epitopes of the protein recognized as antigens in a highly selective manner were predicted using the BepiPred 2.0 program. Overall epitope probability was determined based on the value of the propensity scales, and the probability value of a specific residue greater than a user-defined threshold (by default 0.5) was considered for B-cell linear epitopes (Figure 2). These variable domains might represent unique epitopes. The surface probability plot indicates that most of the amino acids were surface exposed. Further, to assess in vivo antigenic characteristics of the LFliC protein, western blot analysis using mouse anti-LI hyperimmune serum was performed. The serum identified a distinct band near 33 kDa, which shows that mouse anti-LI immune serum had intensive immunoreactivity with the LFliC protein (Figure 3). The presence of antibodies specific to a particular antigen protein indicates that immune response against the protein could be induced in the infected host.
Computational approach to predict conserved B-cell epitopes in the LI0570 protein sequence. A Epitope probability prediction plot of the protein LI0570. B Prediction of B-cell epitopes by different propensity scales. The BepiPred 2.0 program was used for linear epitope prediction of the LFliC protein. The cut-off value (0.5) was automatically determined by the BepiPred 2.0 program.
Western blot analysis of purified LFliC. The protein was strongly recognized by mouse hyperimmune serum obtained from a mouse immunized with L. intracellularis. The arrow in lane A indicates a ~ 33 kDa band, which is the predicted size of LFliC fused with 6×His. Serum from an unimmunized mouse was used as a negative control (lane B). Lane M, size marker.
LFliC characterization
Flagellin, a TLR5 agonist, is a potent activator of the innate immune system. To evaluate the immune stimulatory capacity of LFliC protein, bioactivities of TLR5 and IL-8 were assessed in the presence of purified LI flagellin protein using HEK-Blue™-hTLR5 cells stimulated with the flagellin protein. Increased TLR5 surface expression was detected between 1 and 7 h post-infection in cells stimulated with LFliC (Figure 4). We observed an approximate sixfold increase in the surface expression of TLR5 on cells following 7 h of stimulation compared with that on untreated cells (Figure 4). IL-8 mRNA was also upregulated in pulsed cells (Figure 5). The stimulation led to approximate 590- and 515-fold increases in IL-8 mRNA level in cells treated with LFliC protein at concentrations of 10 and 100 ng/mL, respectively (Figure 5).
Functional characterization of LI0570 protein. Representative FACS histogram of the surface marker-positive cell population. TLR5 expression was measured in HEK-Blue™-hTLR5 cells stimulated with 100 ng/mL of LFliC for various time periods (1, 3, 4, and 7 h). Data shown represent a histogram of samples stained with PE-labeled anti-hTLR5 antibody.
In vitro cytotoxicity assay
In vitro cytotoxicity of the LFliC protein was assessed in the IEC-18 cell line by MTT colorimetric assay following exposure of the LFliC protein for 24 h. After stimulation with the cells at various concentrations of the protein, salt formazan derivatives produced by the mitochondria activity were estimated by absorbance at 560 nm. No significant difference in absorbance value was observed in the cells treated with concentrations of 0.1, 1, 5, and 10 ng/mL LFliC protein (Figure 6). Considering the selective ability of living cells to reduce 3-[4, 5-dimethyl-thiazol-2-yl]-2, 5 diphenyl tetrazolium bromide (MTT) into formazan [19], the rate of conversion to formazan was associated with the metabolic activity of the cells. This result shows that the target protein might not have affected the deteriorative viability of the intestinal cells.
Discussion
Porcine PE caused by L. intracellularis is a common intestinal disease affecting susceptible pigs and poses a substantial economic loss to swine industries. Due to the obligate intracellular life style of the pathogen, conventional laboratory approaches to reveal molecular genetic traits such as antibiotic resistance have been restricted, and development of prevention strategies has been hampered [20]. Despite the completion of LI genome sequencing, protein immunogens against LI infection have yet to be characterized. Proteomic and comparative transcriptional analyses were recently conducted to identify functions related to the pathogenic mechanisms of the LI proteins [7, 21].
In this study, we predicted functional properties and the antigenic potential of the putative LI flagellin protein LI0570. By BLASTP analysis, the amino acid sequence of LI0570 displayed a certain degree of similarity to the amino acid sequence of a flagellin-associated protein in D. alaskensis, E. coli K-12, P. aeruginosa, and C. difficile. Particularly, the B-type flagellin of P. aeruginosa showed 40% similarity of sequence identity to LI0570 and partially contributed to effective production of a protective antibody against P. aeruginosa infection [22]. Immunotherapy with IgG antibodies specific to the N-terminus of the B-type flagellin protein also resulted in effective reduction of mortality and morbidity in a pathogen-infected murine burn model [23]. Given that LI0570 showed substantial similarity to the N-terminal region of P. aeruginosa (Figure 1), the putative LFliC could hypothetically be considered a target for the host immune system.
In this work, the capacity of LFliC as a potential immunogen was confirmed by in silico analysis for linear B-cell epitope prediction and in vitro immunoblotting (Figures 2, 3). Given that an antigen–antibody interaction is fundamental to inducing a humoral immune response to invading pathogens [24], prediction of B-cell epitopes is necessary to evaluate its potential antigenic property. To elucidate whether the predicted B-cell epitope within the LFliC protein reflected its actual antigenicity, western blot analysis was performed using mouse hyperimmune serum raised against LI infection (Figure 3). An immuno-reactive band corresponding to the predicted size of the LFliC protein (33 kDa) was recognized with mouse polyclonal antibodies against LI, which indicated that the LFliC protein may possess potential antigenic determinants. However, the epitope recognition by the mouse anti-LI serum may not guarantee the recognition by a swine convalescent serum, which is still needed to be confirmed in the further study.
Bacterial flagellin, a pathogen-associated molecular pattern (PAMP), binds to TLR5 on the surface of dendritic cells (DC) or epithelial cells [25] and directly or indirectly induces innate and adaptive immune responses [26]. The D1 and D2 domains of bacterial flagellin, two highly conserved regions, are crucial for the recognition of TLR5 [27]. Following activation with the TLR5 ligand flagellin, TLR5-dependent secretion of IL-8, which is controlled by activation of the transcription factor NF-κB and MAPK, necessitates stimulating a signal transduction cascade via the TLR5 pathway [17]. In this study, LFliC protein effectively induced enhanced expression of TLR5 on HEK cells (Figure 4). Further, the transcriptional level of chemokine IL-8 mRNA, which is a TLR5-mediated proinflammatory gene, also increased in the stimulated HEK cells (Figure 5). The results imply that LI0570 possesses intrinsic immuno-stimulatory characteristics involved in inducing innate immunity. Further, cell-mediated immunity is an important feature involved in host defense against intracellular bacterial infection [28]. Given that previous studies reported that bacteria flagellin functions as stimulators for differentiation of CD4+ T cell via DC activation [8], more work is needed to understand the involvement of the LFliC protein in T-cell immunity mediated by DC.
Taken together, the results presented suggest that the LI0570 protein annotated by NCBI as a putative flagellin and related hook-associated protein of LI shows immuno-antigenicity in silico and in vitro, providing evidence that the protein could produce antigen-specific antibodies in vivo. Further, the capacity of LI0570 to induce increased expression of TLR5 suggests that the prominent antigens trigger innate immunity and prime antigen-specific adaptive immunity. Currently, a licensed avirulent live vaccine strain against LI is commercially available and has the potential for reactogenicity [29]. The structural and functional findings of LI0570 in this study could aid in the production of safe recombinant vaccines, and these results will serve as an important groundwork for improving the prevention of PE more generally.
References
Lawson GHK, Gebhart CJ (2000) Proliferative enteropathy. J Comp Pathol 122:77–100
Ohta T, Kimura K, Katsuda K, Kobayashi H, Mikami O, Haritani M, Onodera T (2017) Proliferative enteropathy caused by Lawsonia intracellularis in chickens. J Comp Pathol 156:158–161
Ireton K (2013) Molecular mechanisms of cell–cell spread of intracellular bacterial pathogens. Biol Open 3:130079
Lamason RL, Welch MD (2017) Actin-based motility and cell-to-cell spread of bacterial pathogens. Curr Opin Microbiol 35:48–57
Josenhans C, Suerbaum S (2002) The role of motility as a virulence factor in bacteria. Int J Med Microbiol 291:605–614
Ribet D, Cossart P (2015) How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect 17:173–183
Vannucci FA, Gebhart CJ (2014) Recent advances in understanding the pathogenesis of Lawsonia intracellularis infections. Vet Pathol 51:465–477
Didierlaurent A, Ferrero I, Otten LA, Dubois B, Reinhardt M, Carlsen H, Blomhoff R, Akira S, Kraehenbuhl JP, Sirard JC (2004) Flagellin promotes myeloid differentiation factor 88-dependent development of Th2-type response. J Immunol 172:6922–6930
Huleatt JW, Jacobs AR, Tang J, Desai P, Kopp EB, Huang Y, Song L, Nakaar V, Powell TJ (2007) Vaccination with recombinant fusion proteins incorporating Toll-like receptor ligands induces rapid cellular and humoral immunity. Vaccine 25:763–775
The BLAST Sequence Analysis Tool. http://www.ncbi.nlm.nih.gov/BLAST/. Assessed 10 Dec 2017
The FASTA Search Programs. http://www2.ebi.ac.uk/fasta3/. Assessed 10 Dec 2017
The Multiple Sequence Alignment Program CLUSTALW. http://www2.ebi.ac.uk/clustalw/. Assessed 10 Dec 2017
BepiPred 2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. http://www.cbs.dtu.dk/services/BepiPred/. Assessed 10 Dec 2017
Closter Jespersen M, Peters B, Nielsen M, Marcatili P (2017) BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res 45:W24–W29
Jawale CV, Chaudhari AA, Lee JH (2014) Generation of a safety enhanced Salmonella Gallinarum ghost using antibiotic resistance free plasmid and its potential as an effective inactivated vaccine candidate against fowl typhoid. Vaccine 32:1093–1099
Feng T, Cong Y, Alexander K, Elson CO (2012) Regulation of Toll-like receptor 5 gene expression and function on mucosal dendritic cells. PLoS ONE 7:e35918
Yu Y, Zeng H, Lyons S, Carlson A, Merlin D, Neish AS, Gewirtz AT (2003) TLR5-mediated activation of p38 MAPK regulates epithelial IL-8 expression via posttranscriptional mechanism. Am J Physiol Gastrointest Liver Physiol 285:G282–G290
Won G, Lee JH (2016) Multifaceted immune responses and protective efficacy elicited by a recombinant autolyzed Salmonella expressing FliC flagellar antigen of F18+ Escherichia coli. Vaccine 34:6335–6342
Mosmann T (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63
Jacobson M, Fellström C, Jensen-Waern M (2010) Porcine proliferative enteropathy: an important disease with questions remaining to be solved. Vet J 184:264–268
Watson E, Alberdi MP, Inglis NF, Lainson A, Porter ME, Manson E, Imrie L, Mclean K, Smith DG (2014) Proteomic analysis of Lawsonia intracellularis reveals expression of outer membrane proteins during infection. Vet Microbiol 174:448–455
Ahmadi H, Behrouz B, Irajian G, Amirmozafari N, Naghavi S (2017) Bivalent flagellin immunotherapy protects mice against Pseudomonas aeruginosa infections in both acute pneumonia and burn wound models. Biologicals 46:29–37
Barnea Y, Carmeli Y, Gur E, Kuzmenko B, Gat A, Neville LF, Eren R, Dagan S, Navon-Venezia S (2006) Efficacy of antibodies against the N-terminal of Pseudomonas aeruginosa flagellin for treating infections in a murine burn wound model. Plast Reconstr Surg 117:2284–2291
Slifka MK, Antia R, Whitmire JK, Ahmed R (1998) Humoral immunity due to long-lived plasma cells. Immunity 8:363–372
Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL (2001) Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol 167:1882–1885
McSorley SJ, Ehst BD, Yu Y, Gewirtz AT (2002) Bacterial flagellin is an effective adjuvant for CD4+ T cells in vivo. J Immunol 169:3914–3919
Lu J, Sun PD (2012) The structure of the TLR5-flagellin complex: a new mode of pathogen detection, conserved receptor dimerization for signaling. Sci Signal 5:pe11
Kägi D, Ledermann B, Bürki K, Hengartner H, Zinkernagel RM (1994) CD8+ T cell-mediated protection against an intracellular bacterium by perforin-dependent cytotoxicity. Eur J Immunol 24:3068–3072
Meeusen ENT, Walker J, Peters A, Pastoret PP, Jungersen G (2007) Current status of veterinary vaccines. Clin Microbiol Rev 20:489–510
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GW conducted the experiments; JHL designed the experiments and JHL and GW wrote the paper. Both authors read and approved the final manuscript.
Acknowledgements
This work was supported by the Technological Innovation R&D Program (S2448723) funded by the Small and Medium Business Administration (SMBA, Korea).
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Won, G., Lee, J.H. Antigenic and functional profiles of a Lawsonia intracellularis protein that shows a flagellin-like trait and its immuno-stimulatory assessment. Vet Res 49, 17 (2018). https://doi.org/10.1186/s13567-018-0515-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13567-018-0515-0