Abstract
Interleukin 1 and its receptors are associated with allergic diseases such as asthma. In the present study, we measured DNA methylation at the IL1R1 and IL1R2 gene loci and assessed for associations with asthma-related phenotypes and gene expressions. We found that asthmatic and atopic individuals have higher IL1R2 promoter DNA methylation than control subjects. Additionally, we observed a negative correlation between DNA methylation at the IL1R2 promoter and IL1R2 mRNA expression. These results suggest for the first time that IL1R2 promoter DNA methylation is associated with its gene repression in allergic diseases such as asthma.
Similar content being viewed by others
Introduction
Interleukin 1 (IL1) plays a key role in the inflammatory process of asthma [1]. We reported the association of polymorphisms within the IL1 receptors type I (IL1R1) and type II (IL1R2) gene loci with asthma and atopy in the French Canadian Saguenay–Lac-Saint-Jean (SLSJ) asthma study [2, 3]. The IL1R2 gene expression signature in allergic asthma has also been described [4–6]. Epigenetics has received tremendous attention, and variations in DNA methylation (DNA-Me) in candidate genes have been reported associated with asthma and allergic related disorders [7–12]. These findings underline the relevance of genetic and epigenetic profiling to identify pathways associated with allergic diseases. Such a combined approach will facilitate the understanding of the functional impacts of genetic and epigenetic variations on transcription and molecular mechanisms involved in allergic diseases. In this study, we hypothesized that DNA-Me in the promoters of IL1R1 and IL1R2 is associated with asthma and/or atopy.
Clinical characteristics and methods
Clinical characteristics of the 93 individuals (21 non-atopic asthmatic, 26 atopic asthmatic and 21 atopic individuals, and 25 non-asthmatic non-atopic controls) from the Saguenay–Lac-Saint-Jean asthma familial collection [13] and included in the analysis are shown in Table 1. Ethics committee approved the study, and all subjects gave informed consent. Based on previous genetic [14] and epigenetic analyses [15], methylation at 1 CpG in promoter and 3 CpGs in exon 1 of IL1R1 (Additional file 1: Figure S1) and 5 CpGs in promoter of IL1R2 (Fig. 1a) was measured. DNA-Me differences (Δβ) between affected (individuals with asthma, atopy, or both) and non-affected individuals were assessed and DNA-Me was correlated with gene expression for each CpG. DNA-Me was measured on DNA extracted from blood (Blood and Cell Culture DNA Midi Kit, Qiagen, Canada) using bis-pyrosequencing (EpiTech Bisulfite Kits, Pyromark PCR Kit, Pyromark Gold Q24 Reagents, Qiagen, Canada). PCR primers were designed using PyroMark Assay Design software (v2.0.1.15) (Qiagen,Canada). Total RNA was extracted from whole blood (RNeasy Plus Mini Kit, Qiagen, Canada) using a subset of affected and non-affected individuals (n = 30). For each sample, RNA was converted into cDNA (qScript™ cDNA SuperMix, Quanta Biosciences, USA), and mRNA quantification was determined (PerfeCTa® qPCR FastMix®, Quanta Biosciences, USA) using the two standard curves method with RPLP0 as a reference gene [16].
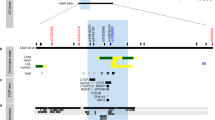
Association between CpGs’ DNA methylation levels for IL1R2 and gene expression in asthma and atopy. a Schematic representation of IL1R2, location of epigenotyped CpG sites, and pairwise correlations between CpG sites. b Mean DNA-Me levels for CpG2 and CpG3-4 of IL1R2 in control and affected subjects (individuals with asthma, atopy, or both). c Correlation between DNA-Me level of IL1R2-CpG2 and mRNA level. d Correlation between mean DNA-Me level of IL1R2-CpG3 and 4 and mRNA level
The association between IL1R1 and IL1R2 DNA-Me levels and asthma and/or atopy at each CpG was analyzed by logistic regression considering age and sex as covariates [9]. Gene expression analysis by phenotype was not performed as control group sample size was insufficient (n = 4). The association between DNA-Me and mRNA levels was assessed by Spearman correlation. CpG dinucleotides with r > 0.6 were combined before they were tested for associations with asthma and/or atopy and for correlation with gene expressions. Δβ with p value < 0.05 was considered statistically significant. Statistical analyses were conducted using the statistical software SPSS (v11.5.0, IBM, USA).
Results
In this study, we detected higher levels of DNA-Me at IL1R2 among affected individuals (i.e., with asthma, atopy, or both) as compared to non-affected controls (Δβ = 8.02 %, p value = 0.013, and Δβ = 3.72 %, p value = 0.012 for IL1R2-CpG2 and the mean for CpG3 and CpG4, respectively (Table 2, Fig. 1b)). Atopic and non-atopic asthma were associated with DNA-Me at IL1R2 but not atopy alone (data not shown). We also observed that DNA-Me at IL1R2-CpG2 was negatively correlated with its mRNA levels (r = −0.511, p value = 0.004) (Fig. 1c), but it was not correlated for CpG3 and CpG4 (Fig. 1d).
Discussion
An epigenetic signature has also been identified for IL1R2 promoter in systemic lupus erythematosus (SLE) [15]. The risk of allergic disorders was significantly increased in SLE patients, which suggests that these conditions share some common biomarkers [17]. The negative correlation we observed between DNA-Me and gene expression levels for IL1R2 may be due to stoichiometry. Methylation may limit access of a transcription factor to DNA and hinders transcriptions [18]. We identified potential binding sites for transcription factors relevant to asthma near the CpG dinucleotide sites of IL1R2 analyzed (Additional file 2: Figure S2) which could explain the inverse correlation between methylation and gene expression [19]. Noteworthy is the potential binding site for nuclear factor kappa B/c-rel (NFKB) at the IL1R2 promoter; it is involved in inflammation through several pathways, including IL1 signalization [20]. Given that IL1R2 acts as a decoy receptor to antagonize the bound ligand [21], our data prompted the speculation that hypermethylation of IL1R2 in asthma and atopy negatively regulates IL1R2 expression and less decoy receptors are available to reduce the downstream pro-inflammatory response of IL1 in the presence of unchanged IL1R1 level [22, 23]. Unlike IL1R1, IL1R2 does not have an intracellular domain and the formation of IL1-IL1R2 complex inactivates the IL1 downstream signaling cascade; hence, silences the role of IL1 in inflammation. Functional study will be needed to investigate the impact of observed epi-variations on the production of expressed receptors. This hypothesis could be attributed to both asthma and atopy as IL1R2 non-signaling receptor is suspected to influence Th2 imbalance [24], and both disorders are driven by Th2 allergic lung inflammation [25, 26].
To our knowledge, this is the first report of (1) a hypermethylation signature of IL1R2 promoter in asthma with or without atopy and (2) an inverse correlation between methylation at IL1R2 promoter and its gene expression. Together, they underline the relevance of IL1R2 as a potential biomarker of asthma and atopy. Further work is needed to understand the interactions between environmental exposures and epigenetic modifications like the ones identified in this study. Such understanding will aid the discovery of disease mechanisms associated and development of more effective therapies.
Abbreviations
- CpG:
-
cytosine-phosphate-guanine
- DNA-Me:
-
DNA methylation
- IL1:
-
interleukin 1
- IL1R1:
-
interleukin 1 receptor type 1
- IL1R2:
-
interleukin 1 receptor type 2
- RPLP0:
-
ribosomal protein, large, P0
- SLE:
-
systemic lupus erythematosus
- SLSJ:
-
Saguenay–Lac-Saint-Jean
- Δβ :
-
difference of methylation
References
Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996;87:2095–147.
Daley D, Lemire M, Akhabir L, Chan-Yeung M, He JQ, McDonald T, et al. Analyses of associations with asthma in four asthma population samples from Canada and Australia. Hum Genet. 2009;125:445–59.
Daley D, Park JE, He JQ, Yan J, Akhabir L, Stefanowicz D, et al. Associations and interactions of genetic polymorphisms in innate immunity genes with early viral infections and susceptibility to asthma and asthma-related phenotypes. J Allergy Clin Immunol. 2012;130:1284–93.
Laprise C, Sladek R, Ponton A, Bernier MC, Hudson TJ, Laviolette M. Functional classes of bronchial mucosa genes that are differentially expressed in asthma. BMC Genomics. 2004;5:21.
Chamberland A, Madore A-M, Tremblay K, Laviolette M, Laprise C. A comparison of two sets of microarray experiments to define allergic asthma expression pattern. Exp Lung Res. 2009;35:399–410.
Pociot F, Molvig J, Wogensen L, Worsaae H, Nerup J. A TaqI polymorphism in the human interleukin-1 beta (IL-1 beta) gene correlates with IL-1 beta secretion in vitro. Eur J Clin Invest. 1992;22:396–402.
Morales E, Bustamante M, Vilahur N, Escaramis G, Montfort M, de Cid R, et al. DNA hypomethylation at ALOX12 is associated with persistent wheezing in childhood. Am J Respir Crit Care Med. 2012;185:937–43.
Reinius LE, Gref A, Saaf A, Acevedo N, Joerink M, Kupczyk M, et al. DNA methylation in the neuropeptide S receptor 1 (NPSR1) promoter in relation to asthma and environmental factors. PLoS One. 2013;8:e53877.
Naumova AK, Al Tuwaijri A, Morin A, Vaillancourt VT, Madore AM, Berlivet S, et al. Sex- and age-dependent DNA methylation at the 17q12-q21 locus associated with childhood asthma. Hum Genet. 2013;132:811–22.
Wang IJ, Karmaus WJ, Chen SL, Holloway JW, Ewart S. Effects of phthalate exposure on asthma may be mediated through alterations in DNA methylation. Clin Epigenetics. 2015;7:27.
Seumois G, Chavez L, Gerasimova A, Lienhard M, Omran N, Kalinke L, et al. Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility. Nat Immunol. 2014;15:777–88.
Yang IV, Pedersen BS, Liu A, O’Connor GT, Teach SJ, Kattan M, et al. DNA methylation and childhood asthma in the inner city. J Allergy Clin Immunol. 2015;136:69–80.
Laprise C. The Saguenay-Lac-Saint-Jean asthma familial collection: the genetics of asthma in a young founder population. Genes Immun. 2014;15:247–55.
Smith AJ, Keen LJ, Billingham MJ, Perry MJ, Elson CJ, Kirwan JR, et al. Extended haplotypes and linkage disequilibrium in the IL1R1-IL1A-IL1B-IL1RN gene cluster: association with knee osteoarthritis. Genes Immun. 2004;5:451–60.
Lin SY, Hsieh SC, Lin YC, Lee CN, Tsai MH, Lai LC, et al. A whole genome methylation analysis of systemic lupus erythematosus: hypomethylation of the IL10 and IL1R2 promoters is associated with disease activity. Genes Immun. 2012;13:214–20.
Wang T, Liang ZA, Sandford AJ, Xiong XY, Yang YY, Ji YL, et al. Selection of suitable housekeeping genes for real-time quantitative PCR in CD4(+) lymphocytes from asthmatics with or without depression. PLoS One. 2012;7:e48367.
Shen TC, Tu CY, Lin CL, Wei CC, Li YF. Increased risk of asthma in patients with systemic lupus erythematosus. Am J Respir Crit Care Med. 2014;189:496–9.
Blattler A, Farnham PJ. Cross-talk between site-specific transcription factors and DNA methylation states. J Biological Chemistry. 2013;288:34287–94.
Turker MS. Gene silencing in mammalian cells and the spread of DNA methylation. Oncogene. 2002;21:5388–93.
Acuner Ozbabacan SE, Gursoy A, Nussinov R, Keskin O. The structural pathway of interleukin 1 (IL-1) initiated signaling reveals mechanisms of oncogenic mutations and SNPs in inflammation and cancer. PLoS Comput Biol. 2014;10:e1003470.
Colotta F, Re F, Muzio M, Bertini R, Polentarutti N, Sironi M, et al. Interleukin-1 type II receptor: a decoy target for IL-1 that is regulated by IL-4. Science. 1993;261:472–5.
Colotta F, Dower SK, Sims JE, Mantovani A. The type II ‘decoy’ receptor: a novel regulatory pathway for interleukin 1. Immunol Today. 1994;15:562–6.
Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39:1003–18.
Sims JE, Gayle MA, Slack JL, Alderson MR, Bird TA, Giri JG, et al. Interleukin 1 signaling occurs exclusively via the type I receptor. Proc Natl Acad Sci U S A. 1993;90:6155–9.
Islam SA, Luster AD. T cell homing to epithelial barriers in allergic disease. Nat Med. 2012;18:705–15.
Paul WE. History of interleukin-4. Cytokine 2015. doi:10.1016/j.cyto.2015.01.038.
Acknowledgements
The authors thank all families for their valuable participation. This study was supported by the Canadian Institutes of Health Research (CIHR) Catalyst Grant: Environments, Genes, and Chronic Disease. Catherine Laprise is the chairholder of the Canada Research Chair in Environment and Genetics of Respiratory Disorders and Allergy and Director of the Asthma Strategic Group of the Respiratory Health Network (RHN) of Fonds de recherche du Québec—Santé (FRQS) and researcher of the AllerGen NCE. Valérie Gagné-Ouellet received a Summer Student Research Training Award from the AllerGen NCE Inc. and a master degree studentship award from the RHN. Anne-Marie Boucher-Lafleur received a Summer Student Research Training Award from the AllerGen NCE Inc. Simon-Pierre Guay is the recipient of a Doctoral Research Award from the CIHR. Luigi Bouchard is a Junior Research Scholar from the FRQS and member of the FRQS-funded Centre de recherche clinique Étienne-Le Bel (affiliated with Centre hospitalier de l’Université de Sherbrooke).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
VGO carried out the pyrosequencing and gene expression studies, performed the statistical analysis, and drafted the manuscript. SPG carried out the pyrosequencing primers design. AMBL participated in the pyrosequencing process. LB participated in the study design and revised the manuscript. CL led each step of the current study including the asthma familial collection build and management, study design, laboratory works, statistical analysis, as well as manuscript redaction and revision. All authors read and approved the final manuscript.
Additional files
Additional file 1: Figure S1.
Schematic representation of IL1R1 and location of epigenotyped CpG sites. This figure illustrates a simplified schematic representation of IL1R1 and location of selected CpG dinucleotide sites and pairwise correlations between each CpG.
Additional file 2: Figure S2.
Potential binding sites for transcription factors in IL1R1 and IL1R2. This figure shows a segment of the primary sequences of IL1R1 and IL1R2. Both sequences lie in the respective gene promoter (UCSC Genome Browser assembly GRCh38/hg38). Epigenotyped CpG sites are shown in blue and are numbered from the distal part of the promoter. Exons are shown in red. Potential binding sites for transcription factors in differentially methylated loci are shown by underlined sequences, and the name of the transcription factors are indicated underneath.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Gagné-Ouellet, V., Guay, SP., Boucher-Lafleur, AM. et al. DNA methylation signature of interleukin 1 receptor type II in asthma. Clin Epigenet 7, 80 (2015). https://doi.org/10.1186/s13148-015-0114-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13148-015-0114-0