Abstract
Background
Libyostrongylus douglassii, Libyostrongylus dentatus and Libyostrongylus magnus are nematodes that infect ostriches. The first species has been identified in ostriches from Africa, Europe, Americas and Oceania. Although the natural range of ostriches is Africa, L. dentatus was first described in birds from the USA and later identified in Brazil, where co-infections with L. douglassii have been commonly reported. Libyostrongylus magnus is known from the original description only. There are a few reports on infections with L. douglassii in ostriches from Africa and all farmed birds examined are from the southern region of the continent. The aim of this report was to verify Libyostrongylus spp. infections in wild ostriches from Ethiopia. Fecal samples from ostriches, Struthio molybdophanes, were collected and submitted to coproculture. Infective larvae were identified to the species level based on general morphology and morphometry. In addition, phylogenetic analysis of the first and second internal transcribed spacer (ITS1 and ITS2) of the nuclear ribosomal DNA was performed.
Results
Infective larvae from Ethiopian ostriches had the morphological characteristics of L. dentatus. Confidence interval estimate for sheath tail length from Ethiopian Libyostrongylus sp. isolates overlapped one for Brazilian L. dentatus. Neighbor-joining and Maximum Likelihood phylogenetic trees based on sequences of the ITS1 and ITS2 regions revealed that the Ethiopian samples belong to the L. dentatus species clade. Monospecific infections with L. dentatus were confirmed in Ethiopian wild ostriches, opposed to the co-infections typically found in the Americas.
Conclusions
To our knowledge, this is the first record of L. dentatus from African ostriches, the region from which this parasite originated.
Similar content being viewed by others
Background
Ostriches, Struthio camelus Linnaeus, 1758 (Aves: Struthionidae), are infected by several species of helminths [1, 2]. In recent years, studies on nematodes of the genus Libyostrongylus, commonly known as wireworms, have intensified [3,4,5,6]. The genus Libyostrongylus comprises Libyostrongylus dentatus Hoberg, Lloyd & Omar, 1995, Libyostrongylus douglassii Lane, 1923 [7] and Libyostrongylus magnus Gilbert, 1937. Species of this genus inhabit the proventriculus and ventriculus of ostriches [4, 8, 9] and feed on blood [1, 10]. Infection with L. douglassii causes a disease known as rotten stomach, responsible for high mortality rates in young, and occasionally, adult birds [10,11,12]. The pathogenic outcomes of infections with L. dentatus [13] and L. magnus are not determined.
Libyostrongylus douglassii has been found in ostriches raised in South Africa [7, 14, 15], Oceania [1, 16], the Americas [4,5,6, 9, 13, 17,18,19,20] and Europe [2, 21,22,23,24,25,26]. Although Africa is the continent of origin of ostriches, L. dentatus has never been reported in this continent. This species was first described in birds raised in the USA [13] and later reported in Brazil [4,5,6, 9, 17, 18]. Due to morphological similarities of L. douglassii and L. dentatus, it was suggested that the latter has been overlooked [9, 13, 18]. Libyostrongylus magnus was described in the Ukraine in ostriches originating from Ethiopia [27, 28]. The females of this species are smaller than males and the ovejector is larger than that in L. douglassii and L. dentatus. No morphological and genetic characterizations of L. magnus infective larvae are available [29] and there are no recent reports of this species.
Although L. douglassii was described from material in South Africa [7], as far as we know, there are only two other published reports of this nematode from ostriches born and maintained in farmed ostriches from Africa. One is the first article on the resistance of this nematode to levamisole [14], and the other reports the parasite in ostriches from the Highveld region of Zimbabwe [15]. There are no reports of Libyostrongylus spp. infections in wild ostriches from South Africa, or from other African countries. This paper reports, for the first time, infections with Libyostrongylus spp. in wild Ethiopian ostriches.
Methods
Samples
Faeces from 5 ostriches (Struthio molybdophanes Reichenow, 1883) were collected at Abijata-Shalla Lakes National Park located in Oromia Regional State, Ethiopia (geographical location: 7°50'N, 38°50'E). Faeces were cultured [30] to obtain infective larvae (L3), at the Laboratory of Veterinary Parasitology, College of Veterinary Medicine and Agriculture, Addis Ababa University, Bishoftu. L3 were preserved in 70% ethanol and shipped to Brazil.
Morphology of infective larvae
Four hundred L3 from each of the 5 hosts were identified under a microscope based on the general morphology of L3, especially the features of the tail, which differentiate L. douglassii from L. dentatus [18]. From this group containing 400 L3, the total length and the sheath tail length was measured for 100 L3 [18]. Images of L3 were captured using a Zeiss Axioplan microscope (Jena, Germany) equipped with differential interference contrast, an AxioCam Mrc5 and the Axiovision program (Germany) at the Universidade Estadual do Norte Fluminense Darcy Ribeiro.
Statistical analysis
Ethiopian Libyostrongylus sp. L3 were investigated in detail by employing statistical analyses using confidence intervals (CI) of measurements of total length and sheath tail length of at least 100 specimens of L. douglassii and L. dentatus from 14 locations in Brazil [5] and of the 100 specimens from Ethiopia that were measured. The L3 larvae of L. douglassii and L. dentatus from Brazil were fixed in 70% ethanol; these were collected in previous studies performed on Brazilian farmed ostriches [5, 6]. The CI of the measurements of L3 were statistically estimated using the following mixed model:
where Yijk is the observation related to the k-th parasite from j-th region of the i-th species. The fixed effects are described in Greek letters including the intercept of the model (μ ). The error term is represented by eijk assumed normal, independent and identically distributed with mean zero and variance σ2. The variance-covariance structure was tested for normality and independence for the random errors according to published recommendations [31]. The model was fitted to data by means of the PROC MIXED procedure of SAS (v.9, SAS System Inc., Cary, NC, USA) [31]. The variance-covariance structure that most likely described the data was the diagonal heterogeneous variances. Point and interval estimates for total L3 size and sheath tail length for L. douglassii and L. dentatus from Brazil and the 500 specimens from Ethiopia were obtained by assuming a 99% confidence level. Results are presented as least squares means and the lower and upper limits of the 99% CI (99% CI) presented in parentheses. Nonetheless, for statistical comparison purposes using the adjusted Tukey-Cramer test, we adopted the type I error rate of 0.02 to avoid spurious positive statistical findings [32].
DNA extraction, PCR amplification and sequencing
A pool of 50 L3 specimens from each of the 5 hosts, in duplicates, was used for DNA extraction in three steps: samples were frozen in liquid nitrogen, submitted to digestion with proteinase K [33] and then a QIamp® DNA mini kit (Qiagen, Hilden, Germany) was used according to the manufacturer's instructions. Samples were observed under an optical microscope to assure the digestion of the cuticles of the larvae. The DNA was quantified in a ND-1000 NanoDrop Spectrophotometer (Thermo Fisher Scientific, Wilmington, USA). The first and the second internal transcribed spacer regions (ITS1 and ITS2) were amplified by PCR using the primers NC5 and NC13 and NC2 and NC13, respectively, as described elsewhere [34, 35]. The PCR reaction volume was 25 μl containing 10 mM Tris-HCl (pH 8.0), 50 mM KCl, 3 mM of MgCl2, 0.2 mM of each dNTP, 20 ρmol of each primer, 1 unit of Platinum® Taq DNA polymerase (Invitrogen, Carlsbad, USA) and 5 μl of extracted DNA. The reactions were subjected to an initial cycle of 5 min at 94 °C, followed by 40 cycles of 94 °C for 40 s, 55 °C for 40 s, 72 °C for 40 s, and 72 °C for 7 min, in a thermal controller Mastercycler ep system (Eppendorf, Hamburg, Germany). PCR products were electrophoresed on 2.0% agarose gels and visualized using GelRed nucleic acid gel stain (Biotium, Hayward, USA). For PCR product purification, a MinElute PCR purification kit (Qiagen) was used according to the manufacturer’s protocol. Amplicons were directly sequenced in both directions using Big Dye Terminator v3.1 Cycle Sequencing Ready reaction kit (Applied Biosystems, Foster City, USA) as recommended by the supplier in a 3100 Automated DNA Sequencer (Applied Biosystems) located on the RPT01A/IOC- Fiocruz sequencing platform.
Sequence analysis
DNA sequences were analyzed, edited and aligned using Pairwise/Blast/NCBI, DNASTAR Lasergene SeqMan v7.0.0, Bio Edit v7.2.5 [36] and ClustalX version 2.0 [37]. ITS1 and ITS2 sequences were submitted to the GenBank database under the accession numbers KY991371- KY991373. The genetic distances and phylogenetic trees were estimated using MEGA version 7 [38]. Genetic distances were calculated using the Kimura 2-parameter (K2P) model [39], complete deletion treatment and standard error estimated (SE) by a bootstrap of 1000 replicates. ITS1 and ITS2 phylogenetic trees were constructed using Neighbor-Joining (NJ) with K2P model [39], and Maximum Likelihood (ML) with Tamura 3-parameter model (T92) [40] plus gamma distribution (G), as selected by the best-fitting model of DNA substitution using the Bayesian information criterion. Complete deletion parameters and 1000 bootstrap replicates were applied.
All ITS1 and ITS2 Libyostrongylus spp. sequences available in GenBank were used in the dataset for phylogenetic analyses. In addition, three sequences of each species of Trichostrongylus spp., Haemonchus spp., Teladorsagia sp., Ostertagia sp., Spiculopteragia sp. and Cooperia spp. (all belonging to the superfamily Trichostrongyloidea) were included in the dataset. Nematodirus battus (Molineoidae) was designated as the outgroup.
Results
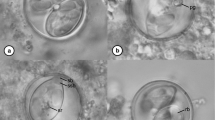
Morphological examination of the L3 samples from Ethiopian ostriches revealed a knob at the tail of larvae and a long and filamentous sheath tail (Fig. 1). L3 with short sheath tail and acute termination were not found in these samples. The statistical analysis of morphometric measurements of L3 revealed that the interaction effect between species and location (αβij) was significant (F-test, residual df = 347, P = 0.002), and the effects were split. Only one comparison between locations based on 99% CI for total length between Brazilian larvae of L. douglassii and L. dentatus was significant, whereas comparisons regarding the sheath tail length of both Brazilian larvae from the 14 Brazilian locations were all significant (Table 1). The 99% CI of the sheath tail length of Libyostrongylus sp. L3 samples obtained from Ethiopia compared to L. dentatus from 14 Brazilian locations were similar in 8 locations and different in the 6 remaining. The total length of the L3 between Ethiopian Libyostrongylus sp. and L. dentatus from Brazil differed in only 5 locations (Table 1).
Although the efficiency of DNA extraction was low, possible due to the poor preservation of the Ethiopian L3 in 70% ethanol, the DNA was successfully extracted from larvae pool of two ostrich samples (2.1 and 4.1). Two partial ITS1 sequences from samples 2.1 and 4.1 (451 and 490 bp, respectively); and one complete ITS2 sequence (237 bp) from 2.1 sample were obtained. The ITS1 alignment of 451 bp showed that the Ethiopian Libyostrongylus sp. sequences were identical. Ethiopian Libyostrongylus sp. sequences also had an insertion of 26 bp when compared to Brazilian L. douglassii ITS1 sequence, as demonstrated previously [3].
ITS1 genetic distance between Libyostrongylus sp. from Ethiopia and Brazilian L. dentatus was 0.003 (SE = 0.002), while between Libyostrongylus sp. from Ethiopia and Brazilian L. douglassii was 0.090 (SE = 0.017), which was the same genetic distance between L. dentatus and L. douglassii from Brazil.
ITS2 genetic distance between Libyostrongylus sp. from Ethiopia and Brazilian L. dentatus was 0.002 (SE = 0.002), while between Brazilian L. douglassii was 0.073 (SE = 0.020), which was the same genetic distance between Brazilian L. dentatus and L. douglassii. A higher value of genetic distance (0.100; SE = 0.025) was obtained when comparing ITS2 sequences from Ethiopian samples with a Libyostrongylus sp. sample of gorilla faeces from Cameroon (JX159958) [41], which coincides with the value obtained between the gorilla Libyostrongylus sp. and L. dentatus from Brazil. In addition, higher values of genetic distance were obtained when comparing L. dentatus and L. douglassii from Brazil (0.073; SE = 0.020).
NJ and ML trees based on the ITS1 dataset showed that both Libyostrongylus sp. sequences from Ethiopia clustered with L. dentatus from Brazil in a monophyletic clade with maximum bootstrap values (NJ and ML = 100%) (Fig. 2), while L. douglassii clustered in another monophyletic clade with high bootstrap values (ML = 99%, NJ = 100%). All Libyostrongylus spp. sequences formed a well-supported clade (ML = 88%, NJ = 90%). In addition, genus-specific clades with strong support were detected for Trichostrongylus spp., Haemonchus spp., Teladorsagia circumcincta, Ostertagia leptospicularis, Spiculopteragia houdemeri and Cooperia spp. (ML = 92–100%, NJ = 97–100%) (Fig. 2).
Maximum Likelihood (ML) phylogenetic tree inferred from ITS1 sequences for Ethiopian Libyostrongylus sp. (this study) and representative GenBank sequences for species of related families (Trichostrongylidae, Cooperiidae). Bootstrap values (> 50%) for nodal support correspond to ML analysis using T92 + G model (italic and bold font) and NJ analysis with K2P model (regular font). Libyostrongylus dentatus clade is indicated in red; filled diamonds correspond to the newly generated sequences for Ethiopian Libyostrongylus sp.
Similarly, the ITS2 ML and NJ trees revealed that Libyostrongylus sp. from Ethiopia grouped with L. dentatus in a monophyletic clade with high bootstrap support (ML and NJ = 99%) (Fig. 3). Libyostrongylus douglassii sequences also grouped in a monophyletic clade but with low bootstrap support (ML = 55%, NJ = 63%), showing a strongly-supported subgroup comprising only Brazilian L. douglassii (ML = 97%). The Libyostrongylus sp. sequence from gorilla (GenBank: JX159958) was resolved as basal to Libyostrongylus spp. clade (Fig. 3), but still clustered in the genus-level clade, with strong and moderate support (ML = 98%, NJ = 87%). Furthermore, genus-specific clusters with strong support were detected for Trichostrongylus spp., Spiculopteragia houdemeri, Cooperia spp., Haemonchus spp., Teladorsagia circumcincta and Ostertagia leptospicularis (ML = 97–100%, NJ = 81–100%).
Maximum Likelihood (ML) phylogenetic tree inferred from ITS2 sequences for Ethiopian Libyostrongylus sp. (this study) and representative GenBank sequences for species of related families (Trichostrongylidae, Cooperiidae). Bootstrap values (> 50%) for nodal support correspond to ML analysis using T92 + G model (italic and bold font) and NJ analysis with K2P model (regular font). Libyostrongylus dentatus clade is indicated in red: the filled diamond corresponds to the newly generated sequence for Ethiopian Libyostrongylus sp.
Discussion
Libyostrongylus spp. infection in ostriches is widely distributed in the world, being a major problem for commercial production of these animals. There is relative lack of data concerning this infection in African countries and none in wild ostriches. In this study we examined Libyostrongylus spp. parasitism in wild ostriches from Ethiopia. Morphological and morphometric data for L3 larvae demonstrated the presence of only one species of Libyostrongylus. i.e. L. dentatus, in these ostriches that was supported by genetic characterization.
The total length of the L3 is a characteristic that helps the differentiation of L. douglassii from L. dentatus but presents an elevated standard deviation that may lead to error [18]. This was further demonstrated in samples from different Brazilian locations [5] in a two-year systematic sampling [6], clearly indicating that sheath tail length is the best morphological parameter to distinguish these species. The statistical analysis presented herein also confirmed that sheath tail length is the best morphological character to discriminate between L. douglassii and L. dentatus.
The observation of the knob at the posterior end of the L3 from Ethiopia clearly indicates that this material belongs to the genus Libyostrongylus [1, 18]. The presence of the long filamentous sheath tail found in Ethiopian samples indicates that the species observed was L. dentatus. This was confirmed by statistical analysis of the sheath tail length that showed a greater similarity between Ethiopian Libyostrongylus sp. and Brazilian L. dentatus than Brazilian L. douglassii, further suggesting that the Ethiopian samples belong to L. dentatus.
Molecular data were used to confirm morphological data. Libyostrongylus sp. sequences from the Ethiopian hosts clustered with L. dentatus sequences from Brazil. Ethiopian samples presented a 26 bp insertion in the ITS1 region that is present only in L. dentatus [3]. Phylogenetic trees based on ITS1 and ITS2 datasets showed a monophyletic group comprising Libyostrongylus sp. from Ethiopia and L. dentatus, with high bootstrap support. These robust phylogenetic results, together with the morphological data, confirm that Libyostrongylus sp. from Ethiopia was L. dentatus and not another known or new Libyostrongylus species.
An ITS2 Libyostrongylus sp. sequence of gorilla faeces from Cameroon [41] clustered with strong support with Libyostrongylus spp. in the present study. Hamad et al. [41] suggested that the detection of Libyostrongylus sp. in gorilla fecal samples was probably due to environmental contamination or consumption of food contaminated by larvae from an ostrich. Although the ITS2 sequence from gorilla faeces belonged to the genus Libyostrongylus in our analysis, high genetic distances relative to both L. douglassii and L. dentatus were found, not allowing the identification of the species. An explanation is that the gorilla fecal sample sequence could represent a species of the genus Paralibyostrongylus, which is closely related to the genus Libyostrongylus. Paralibyostrongylus was regarded as a synonym of Libyostrongylus by Chabaud [42]. However, other authors have considered Libyostrongylus and Paralibyostrongylus as distinct genera that belong to the same subfamily, Libyostrongilinae [43]. Gorillas are the hosts of Paralibyostrongylus hebrenicutus and P. kalinae [44,45,46]; no molecular data for these species are available. Another possibility is that the sequence (JX159958) is another Libyostrongylus species not yet morphologically characterized. Further molecular analysis of Paralibyostrongylus spp. is necessary to verify if the gorilla fecal sample sequence belongs to the genus Paralibyostrongylus or represents another Libyostrongylus species.
Libyostrongylus magnus was described in ostriches from Ethiopia [13, 27, 28]. However, the morphological and genetic characterization presented in our study identified L. dentatus from the Ethiopian samples. The absence of L. magnus in these samples may be explained by the host subspecies studied. We identified L. dentatus in S. molybdophanes, while L. magnus was described in S. c. camelus [13]. The distribution of the two species may possibly vary according to the ostrich subspecies as proposed previously [13].
To our knowledge, this is the first record of L. dentatus outside the Americas, and as a monospecific infection. In our previous studies on farmed ostriches in Brazil, monospecific Libyostrongylus infection was never detected [3, 6]. A population dynamics study of L. dentatus and L. douglassii was performed for two years, analyzing about 192,000 L3 from 40 ostriches, and a monospecific Libyostrongylus sp. infection was never recorded in those birds. The co-infection condition (L. dentatus + L. douglassii) was always found, with a general predominance of L. douglassii [6]. In addition, ostriches from 13 Brazilian farms from distinct regions of the country were also examined and, again, a monospecific infection by Libyostrogylus sp. was never recorded [3]. Furthermore, in both studies, the species that dominates the co-infection was L. douglassii, not L. dentatus. In the present study, only L. dentatus was found in the 2000 L3 examined, indicating that this is the first record of a monospecific Libyostrongylus sp. infection ever reported. More ostriches from Ethiopia, and from distinct African regions, need to be sampled to understand the infection patterns of Libyostrongylus spp.
Conclusions
To our knowledge, this is the first record of L. dentatus infecting ostriches outside the Americas. It is also the first report of L. dentatus infection in wild ostriches from an Ethiopian region. The study demonstrates a scenario of monospecific infections with L. dentatus in Ethiopian wild ostriches, contrary to the typical observation of co-infections in farmed ostriches from the Americas.
Abbreviations
- CI:
-
Confidence intervals
- G:
-
Gamma distribution
- ITS1:
-
First internal transcribed spacer
- ITS2:
-
Second internal transcribed spacer
- L3:
-
Infective third-stage larvae
- ML:
-
Maximum likelihood
- NJ:
-
Neighbor-joining
- SE:
-
Standard error
References
Barton N, Seward D. Detection of Libyostrongylus douglassi in ostriches in Australia. Aust Vet J. 1993;70:31–2.
Ponce Gordo F, Herrera S, Castro A, García Durán B, Martínez Díaz RM. Parasites from farmed ostriches (Struthio camelus) and rheas (Rhea americana) in Europe. Vet Parasitol. 2002;107:137–60.
Andrade JG, Iñiguez AM, Souza AN, Marques VC, de Souza Filho GA, Santos CP, DaMatta RA. Genetic characterization of the blood-sucking nematodes Libyostrongylus dentatus and Libyostrongylus douglassii supports their different evolutionary history. Vet Parasitol. 2013;193:193–9.
Andrade JG, Carvalho ECQ, de Santos CP, DaMatta RA. Mixed infection with Libyostrongylus dentatus and Libyostrongylus douglassii induces a heterophilic inflammatory infiltrate in the proventriculus of ostriches. Avian Pathol. 2011;40:367–70.
Andrade JG, Lelis RT, DaMatta RA, Santos CP. Occurrence of nematodes and anthelmintic management of ostrich farms from different Brazilian states: Libyostrongylus douglassii dominates mixed infections. Vet Parasitol. 2011;178:129–33.
Lelis RT, Andrade JG, Vieira RAM, DaMatta RA, Santos CP. Population dynamics of Libyostrongylus dentatus and L. douglassii of ostriches raised in the northern Rio de Janeiro State, Brazil. Vet Parasitol. 2014;200:147–52.
Cobbold TS. New Entozoon from the ostrich. Zool J Linnean Soc. 1882;16:184–8.
Ederli N, Oliveira F. Differential localization of Libyostrongylus douglassii (Cobbold, 1882) Lane, 1923 and L. dentatus Hoberg, Lloyd, and Omar, 1995 (Nematoda: Trichostrongylidae) in ostrich (Struthio camelus Linnaeus, 1758) proventriculi. J Parasitol. 2009;95:757–9.
Ederli NB, Bonadiman SF, de Moraes Neto AHA, DaMatta RA, Santos CP. Mixed infection by Libyostrongylus douglassii and L. dentatus (Nematoda: Trichostrongylidae) in Struthio camelus (Ratites: Struthioniformes) from Brazil with further morphological characterization of adults. Vet Parasitol. 2008;151:227–32.
Reinecke RK. Veterinary helminthology. Durban: Butterworths; 1983.
Jansson D. Strutsar och andra ratiter, del 2 Enkätundersökning av svenska ratitfarmer 1999. Svensk Veterinärtidning. 2000;11:569–73.
Nel C. Dosing of ostriches for internal parasites. Elsenburg J. 1980;4:31–3.
Hoberg EP, Lloyd S, Omar H. Libyostrongylus dentatus n. sp. (Nematoda: Trichostrongylidae) from ostriches in North America, with comments on the genera Libyostrongylus and Paralibyostrongylus. J Parasitol. 1995;81:85–93.
Malan F, Gruss B, Roper N, Ashburner A, Du Plessis C. Resistance of Libyostrongylus douglassi in ostriches to levamisole. J S Afr Vet Assoc. 1988;59:202–3.
Mukaratirwa S, Cindzi Z, Maononga D. Prevalence of Libyostrongylus douglassii in commercially reared ostriches in the highveld region of Zimbabwe. J Helminthol. 2004;78:333–6.
McKenna P. Libyostrongylus infections in ostriches - a brief review with particular reference to their detection in New Zealand. N Z Vet J. 2005;53:267–70.
Souza LP, Lelis RT, Granja IRA, DaMatta RA, Santos CP. Efficacy of albendazole and moxidectin and resistance to ivermectin against Libyostrongylus douglassii and Libyostrongylus dentatus in ostriches. Vet Parasitol. 2012;189:387–9.
Ederli NB, Oliveira FCR, Lopes CWG, Damatta RA, Santos CP, Rodrigues MLA. Morphological diagnosis of infective larvae of Libyostrongylus douglassii (Cobbold, 1882) Lane, 1923 and L. dentatus Hoberg, Lloyd and Omar, 1995 (Nematoda: Trichostrongylidae) of ostriches. Vet Parasitol. 2008;155:323–7.
Lozada H, Rincón J, López V. Parásitos en avestruces en el Departamento del Meta. Rev Sist Prod Agroecol. 2011;2:34–46.
Mariño-González GA, Ramírez-Hernández A, Cortés-Vecino JA. Libyostrongylus douglassii (Strongylida: Trichostrongylidae) in ostrich (Struthio camelus) farms from Colombia. Vet Parasitol. 2017;235:53–6.
Landman W, Bronneberg R. Libyostrongylus douglassii in ostriches (Struthio camelus ssp.) in the Netherlands: case report and review. Tijdschr Diergeneeskd. 2001;126:484–7. (In Dutch)
Jansson D, Christensson D, Christensson B. Winter survival in Sweden of L 3-stage larvae of the ostrich wireworm Libyostrongylus douglassii. Vet Parasitol. 2002;106:69–74.
Ocal N, Karahan S, Atmaca T. Proliferative response by the ostrich proventriculus in idiopathic gastric stasis: A case report. Acta Vet Hung. 2006;54:213–20.
Yaman M, Durgut R. Parasitic infestations in ostriches and therapy. Turkiye Parazitol Derg. 2005;29:103. (In Turkish)
Schulze C, Grossmann E, Krone O. Case report: Libyostrongylus douglassii-associated proventriculitis in ostriches in Germany. Dtsch Tierarztl Wochenschr. 2006;113:240–2.
Tišljar M, Beck R, Cooper R, Marinculić A, Tudja M, Lukač-Novak I, et al. First finding of libyostrongylosis in farm-reared ostriches (Struthio camelus) in Croatia: Unusual histopathological finding in the brain of two ostriches, naturally infected with Libyostrongylus douglasi. Vet Parasitol. 2007;147:118–24.
Gilbert L. A new nematode, Libyostrongylus magnus n. sp. parasitic in an African ostrich. In: Skrjabin KJ, editor. Papers on helminthology, 30 year jubilee. Moscow: Lenin Academy of Agricultural Science; 1937. p. 180–2.
Huchzermeyer FW. Diseases of farmed crocodiles and ostriches. Rev Sci Tech. 2002;21:265–76.
Ederli NB, Oliveira FCR. Comparative morphology of the species of Libyostrongylus and Codiostomum, parasites from ostriches, Struthio camelus, with a identification key to the species. Rev Bras Parasitol Vet. 2014;23:291–300.
Bonadiman SF, Ederli NB, Soares AKP, Moraes Neto AHA, Santos CP, DaMatta RA. Occurrence of Libyostrongylus sp.(Nematoda) in ostriches (Struthio camelus Linnaeus, 1758) from the north region of the state of Rio de Janeiro, Brazil. Vet Parasitol. 2006;137:175–9.
Vieira RAM, Campos PRSS, da Silva JFC, Tedeschi LO, Tamy WP. Heterogeneity of the digestible insoluble fiber of selected forages in situ. Anim Feed Sci Technol. 2012;171:154–66.
Johnson VE. Revised standards for statistical evidence. Proc Natl Acad Sci USA. 2013;110:19313–7.
Iñiguez AM, Araújo A, Ferreira LF, Vicente ACP. Analysis of ancient DNA from coprolites: a perspective with random amplified polymorphic DNA-polymerase chain reaction approach. Mem Inst Oswaldo Cruz. 2003;98:63–5.
Gasser RB, Chilton NB, Hoste H, Beveridge I. Rapid sequencing of rDNA from single worms and eggs of parasitic helminths. Nucleic Acids Res. 1993;21:2525–6.
Newton L, Chilton N, Beveridge I, Hoste H, Nansen P, Gasser R. Genetic markers for strongylid nematodes of livestock defined by PCR-based restriction analysis of spacer rDNA. Acta Trop. 1998;69:1–15.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser (Oxf). 1999;41:95–8.
Larkin MA, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8.
Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20.
Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol. 1992;9:678–87.
Hamad I, Keita MB, Peeters M, Delaporte E, Raoult D, Bittar F. Pathogenic eukaryotes in gut microbiota of western lowland gorillas as revealed by molecular survey. Sci Rep. 2014;4:6417.
Chabaud AG. Remarques sur la systematique des nematodes Trichostrongyloidea. Bull Soc Zool Fr. 1959;84:473–83.
Gibbons LM, Khalil L. A key for the identification of genera of the nematode family Trichostrongylidae Leiper, 1912. J Helminthol. 1982;56:185–233.
Lane C. Some Strongylata. Parasitology. 1923;15:348–64.
Durette-Desset M-C, Chabaud A, Ashford R, Butynski T, Reid G. Two new species of the Trichostrongylidae (Nematoda: Trichostrongyloidea), parasitic in Gorilla gorilla beringei in Uganda. Syst Parasitol. 1992;23:159.
Rothman J, Bowman D. A review of the endoparasites of mountain gorillas. Companion and exotic animal parasitology. Ithaca, NY: International Veterinary Information Service; 2003.
Acknowledgments
The authors would like to thank Andrèa Carvalho César for proofreading the manuscript.
Funding
This study was funded by the Brazilian fostering agencies Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (AMI 307932/2014-1; RAD 307014/2015-0), Fundação Carlos Chagas Filho do Rio de Janeiro (FAPERJ) (AMI CNE E-26/202.945/2016; JGA E-26/103.390/2012; RAD E-26/202.937/2017) and Fundação de Coordenação de Pessoal de Nível Superior (CAPES).
Availability of data and materials
ITS1 and ITS2 sequences generated and analysed during the current study are available in the GenBank database (KY991371-KY991373).
Author information
Authors and Affiliations
Contributions
JGA completed the morphological, morphometry and phylogenetic characterization. BK and DA collected the samples and performed the coproculture. RAMV, CPS and RAD completed the statistical analysis of the morphometry. CPS completed the morphological characterization. AMI completed the genetic and phylogenetic analysis. CPS and RAD conceived the study; all authors interpreted the data and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Andrade, J.G., Kumsa, B., Ayana, D. et al. First record of the nematode Libyostrongylus dentatus Hoberg, Lloyd & Omar, 1995 (Trichostrongylidae) in ostriches (Struthio camelus Linnaeus, 1758) (Struthionidae) outside the Americas. Parasites Vectors 11, 243 (2018). https://doi.org/10.1186/s13071-018-2815-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-018-2815-7