Abstract
Background
Anaplasma phagocytophilum is wide spread throughout the world and impacts both human and animal health. Several distinct ecological clusters and ecotypes of the agent have been established on the basis of various genetic loci. However, information on the genetic variability of A. phagocytophilum isolates in China represents a gap in knowledge. The objective of this study was to determine the prevalence and genetic characterization of A. phagocytophilum in small ruminants in central and southeastern China.
Methods
The presence of A. phagocytophilum was determined in 421 blood samples collected from small ruminants by PCR. Positive samples were genetically characterized based on 16S rRNA and groEL genes. Statistical analyses were conducted to identify ecotypes of A. phagocytophilum strains, to assess their host range and zoonotic potential.
Results
Out of 421 sampled small ruminants, 106 (25.2%) were positive for A. phagocytophilum. The positive rate was higher in sheep (35.1%, 40/114) than in goats (26.4%, 66/307) (P < 0.05). Sequence analyses revealed that the isolates identified in this study were placed on two separate clades, indicating that two 16S rRNA variants of A. phagocytophilum were circulating in small ruminants in China. However, analysis using obtained groEL sequences in this study formed one cluster, which was separate from other known ecotypes reported in Europe. In addition, a novel Anaplasma sp. was identified and closely related to an isolate previously reported in Hyalomma asiaticum, which clustered independently from all recognized Anaplasma species.
Conclusions
A molecular survey of A. phagocytophilum was conducted in sheep and goats from ten provinces in central and southeastern China. Two 16S rRNA variants and a new ecotype of A. phagocytophilum were identified in small ruminants in China. Moreover, a potential novel Anaplasma species was reported in goats. Our findings provide additional information on the complexity of A. phagocytophilum in terms of genetic diversity in China.
Similar content being viewed by others
Background
Anaplasma phagocytophilum is an obligate intracellular bacterium that replicates mainly in neutrophils of humans and animals [1]. It is thought to be maintained primarily in an enzootic cycle between Ixodes ticks and vertebrate hosts and may cause human, canine, and equine granulocytic anaplasmosis and tick-borne fever in ruminants [2]. In addition, A. phagocytophilum infection has been reported in a variety of wild and domestic animals, especially in rodents and wild deer [3]. In China, the first suspected human case was described in Anhui Province in 2006 [4]; since then, an increasing number of HGA cases have been recorded in six cities and provinces [5].
The survival and replication of A. phagocytophilum in different tick species and a wide range of vertebrate hosts may lead to high genetic variability of the agent, as revealed by analysis of various genetic loci, including 16S rRNA, heat-shock operon (groEL), ankA and genes of major surface proteins [3, 6–10]. Recently, the A. phagocytophilum variants characterized based on the 16S rRNA gene have been typed extensively on the basis of the groEL gene in ticks and animals, and several lineages and ecotypes with different pathogenic abilities were reported in both European countries and USA [3, 8, 11, 12]. The molecular characterization of A. phagocytophilum strains in China was also reported, and several 16S rRNA variants were identified in ticks, small mammals and ungulates [5, 13–16]. However, it is unclear at this stage how the Chinese A. phagocytophilum isolates segregate into lineages or ecotypes and whether these isolates genetically differ from those pathogenic strains reported in Europe and in USA. In this study, the presence of A. phagocytophilum in small ruminants was investigated. The isolates of A. phagocytophilum were characterized by sequence analysis of the 16S rRNA and groEL genes and compared them to the strains reported earlier.
Methods
Study sites and collection of specimens
The survey was performed from March to September during the peak season of tick activities between 2011 and 2015 in rural areas of ten provinces containing 17 counties in central and southeastern China (Table 2). Two to three sites were selected for sampling in each county. EDTA whole-blood samples were taken randomly from the jugular vein of 421 asymptomatic small ruminants (sheep, n = 114; goats, n = 307) and collected in a sterile tube. Total DNA was extracted from 300 μl of blood using the Gentra Puregene Blood Kit (Qiagen, Beijing, China) according to the manufacturer’s instructions.
PCR reactions
Anaplasma phagocytophilum DNA was detected by nested PCR for the amplification of the 16S rRNA gene as previously described [17, 18]. Anaplasma and Ehrlichia genus-specific primers EE1 and EE2 were used for primary reactions, and species-specific primers SSAP2f and SSAP2r were used for nested reactions (Table 1). The length of the nested PCR fragments was 641 base pairs. Positive samples were selected randomly and used for the amplification of the groEL gene [19]. The partial groEL gene sequences (574 bp) were amplified using EphplgroEL(569)F and EphplgroEL(1193)R primers for primary reactions and EphplgroEL(569)F and EphgroEL(1142)R primers for semi-nested reactions (Table 1) [19]. The reaction was performed in an automatic thermocycler (Bio-Rad) with a total volume of 25 μl containing 2.5 μl of 10× PCR buffer (Mg2+ Plus), 2.0 μl of each dNTP at 2.5 mM, 1.25 U of Taq DNA polymerase (TaKaRa, Dalian, China), 2.0 μl of template DNA, 1.0 μl of each primer (10 pmol), and 16.25 μl of distilled water. Genomic DNA extracted from the whole blood of sheep infected with A. phagocytophilum (GenBank accession no. JN558811) was used as the positive control, and sterile water was used as the blank control for each run. Cycling conditions for PCR amplification were: 4 min of denaturation at 94 °C, 35 cycles at 94 °C for 1 min, annealing for 45 s (annealing temperatures of primers are listed in Table 1), and 72 °C for 45 s- 1.5 min (dependent on the target genes), with a final extension step at 72 °C for 10 min. Amplified fragments were run on a 1.0% agarose gel following electrophoresis, staining with ethidium bromide, and then visualized by UV transillumination.
DNA sequencing and phylogenetic analysis
The amplified PCR products were purified using the TaKaRa Agarose Gel DNA Purification Kit Ver.2.0 (TaKaRa), ligated into pGEM-T Easy vector (Promega, Madison, WI, USA) and transformed into Escherichia coli JM109 competent cells (TaKaRa). Two recombinant clones were selected for sequencing using BigDye Terminator Mix (Sangon, Shanghai, China). The obtained sequences were analysed by a BLASTn search in GenBank or by using the ClustalW method in the MegAlign software (DNAStar, Madison, WI, USA). Phylogenetic trees were constructed based on the sequence distance method using the neighbor-joining (NJ) algorithm with the Kimura two-parameter model of the Mega 4.0 software [20].
Statistical analysis
Statistical analysis was conducted using a Chi-square test in Predictive for Analytics Software Statistics 18 (PASW, SPSS Inc., Chicago, IL, USA). P-values of 0.05 or less were considered statistically significant.
Nucleotide sequence accession numbers
The representative sequences obtained in this study have been submitted and deposited in the GenBank database with accession numbers KX272641–KX272643 for 16S rRNA and KX276166–KX276167 for groEL partial sequences.
Results
Out of 421 sampled animals, 106 (25.2%) were positive for A. phagocytophilum (Table 2). The positive rates for A. phagocytophilum at different sampling sites varied from 0 to 75%. The positive rate was higher in sheep (35.1%, 40/114) than in goats (26.4%, 66/307) (χ 2 = 11.090, df = 1, P < 0.05). As shown in Table 2, the agent was detected in fourteen out of seventeen study sites in central and southeastern China.
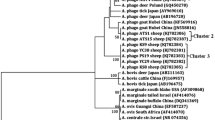
The molecular characterization of A. phagocytophilum isolates in sheep and goats was analysed based on 16S rRNA and groEL genes. Forty-nine 16S rRNA sequences of A. phagocytophilum representative of different geographical locations were obtained in this study. The similarity among 16S rRNA gene sequences ranged from 99.7 to 100%. Phylogenetic analysis revealed that the A. phagocytophilum isolates identified in this study are placed in two separate clades (Fig. 1). Group 1, which contained 45 sequences (GenBank accession no. KX272641), was identical to the following strains: giraffe 2013-6, HB-C3, s4 and sika28, which were detected in the South African giraffe, Haemaphysalis longicornis, cattle and Cervus Nippon (GenBank accession nos. KU870667, KF569915, KX115422 and LC060987), respectively. Group 2 contained four sequences (GenBank accession nos. KX272642) and had 100% identity to the ApGGo2, Ap-SHAX31 and EKY155 strains derived from goats (GenBank accession nos. KM285227, KU321299 and JF807994), ATS1 from sheep (GenBank accession no. KJ782386) and JC3-3 from Procapra gutturosa (GenBank accession no. KM186948).
Twenty-two groEL sequences obtained from the 16S rRNA gene positive samples were identical to each other and showed 96.1% identity to strain HB-MC-A25 of A. phagocytophilum from cattle (GenBank accession no. KF569919). Phylogenetic analysis was conducted with the groEL sequences in this study and sequences identified in different hosts and countries deposited in GenBank (groEL sequences are available in Additional file 1: Table S1). Five major A. phagocytophilum clusters could be proposed (labelled I-V in Fig. 2). The groEL sequences in this study formed one cluster (V), which was separate from the other four clusters or ecotypes reported in Europe. In addition, one isolate (20-5a) identified in a goat was different from known Anaplasma species (Figs. 1 and 2). The 16S rRNA and groEL sequences (GenBank accession nos. KX272643 and KX276167) showed 99.8 and 92.6% identity to the strains BL102-7 of an unclassified Anaplasma species from Hyalomma asiaticum ticks, respectively (GenBank accession nos. KJ410249 and KJ410302). The isolate was closely related to a novel Anaplasma sp. and clustered independently from all known Anaplasma species (Figs. 1 and 2).
Discussion
Anaplasma phagocytophilum is an emerging tick-borne pathogen with veterinary and public health significance [1]. Since the first description of the agent in 1932, many wild and domestic animals have been considered as the reservoir hosts for A. phagocytophilum [1, 21]. In China, this species has been reported in sheep, goats, cattle, deer, yaks, dogs, rabbits and rodents in several provinces, and the infection rates vary in different hosts and/or geographical locations [14–16, 22, 23]. In this study, a molecular survey of A. phagocytophilum was conducted in sheep and goats in ten provinces in central and southeastern China. The positive rates of A. phagocytophilum in sheep and goats (25.2%, 106/421) was higher than those in goats from Jilin (5.7%, 8/35), Henan (13.0%, 6/46) and Hubei (14.5%, 10/69), and were lower than in sheep and goats (40.0%, 56/140) conducted in Gansu Province [13, 14, 16]. However, it is almost comparable with 26.69% prevalence in goats (126/472) shown in a previous report [24]. The positive rates for A. phagocytophilum were variable in sheep and goats as well as different sampling sites in China, and this may be related to sampling time and methods, tick vectors and methodical approaches. Anaplasma phagocytophilum infection was found in 14 of 17 study sites in the present study, implying an extensive geographical distribution of the agent in China. It is well known that A. phagocytophilum could cause a persistent infection in sheep and rodents, which allows them to be reservoirs of infection [25]. The sheep and goats serve as reservoir hosts may facilitate further spread of infection.
Anaplasma phagocytophilum exhibits a high degree of genetic diversity, host tropisms and variation in pathogenicity [7]. Genetic diversity of the agent has been reported in different hosts or geographical locations based on groEL, ankA and msp4 [8–10]. Although high similarities of the 16S rRNA gene sequences were reported, several 16S rRNA gene variants have been identified [17, 26]. In this study, two 16S rRNA variants of A. phagocytophilum were identified in sheep and goats. Phylogenetic analysis revealed that variant 1 (GenBank accession no. KX272641) clustered, with 100% identity, together with the strains isolated from cattle, giraffe, goats and H. longicornis in China, sika deer (Cervus nippon) and cattle in Japan (Fig. 1), indicating that it circulates in a wide range of vertebrate hosts in Asia. Variant 2 (GenBank accession no. KX272642) has been detected in sheep, goats and Mongolian gazelle (Procapra gutturosa) in China and goats in Tunisia and Turkey (Fig. 1), implying that it is distributed worldwide. Moreover, variant 1 (91.8%) appears to be the most dominant strain in sheep and goats in China.
In accordance with the existing genetic diversity at the level of nucleotide sequences of the groEL gene, it has been used for the distinction of lineages or ecotypes represented by a variety of isolates in the environment and clinical samples [9, 11, 26]. A previous report has revealed that A. phagocytophilum circulates in Eastern Europe and belongs to two groESL lineages and the strains pathogenic for humans and animals were clustered within lineage one [26]. Recently, four geographically dispersed ecotypes with different host ranges and zoonotic potential were identified based on groESL in Europe [3]. In the present study, the groEL sequences of A. phagocytophilum in sheep and goats formed one cluster (ecotype V), which was distinct from the known ecotypes in Europe (Fig. 2). According to the previous reports, A. phagocytophilum strains within ecotype I had a wide host range and were commonly isolated from wild and domestic animals as well as humans [3]. Strains that are pathogenic to humans and animals were found within ecotype I, indicating the pathogenic potential of members in this ecotype. Ecotype II was mainly found in roe deer and rodents, ecotype III in rodents and ecotype IV in birds (Fig. 2) [3]. Clinical signs in wild species are very difficult to observe. Little is known about pathogenicity of the different variants of A. phagocytophilum in these species. Ecotype V has been found not only in sheep and goats in this study, but also in ticks and cattle in previous reports (Figs. 1 and 2) [27], suggesting the circulation and wide distribution of this ecotype in China. However, very limited clinical data are available so far, and it is unknown whether the detected strains in ecotype V are pathogenic to humans or livestock animals in China. Clearly, the zoonotic potential of the members within ecotype V need to be further investigated.
Currently, the recognized Anaplasma species include A. phagocytophilum, A. marginale, A. centrale (A. marginale subsp. centrale), A. ovis, A. bovis and A. platys. In addition to the above mentioned species, a novel Anaplasma species designated “Anaplasma capra” has been identified in goats, ticks and humans in northern China [28]. In addition to well-characterized Anaplasma species, a number of new Anaplasma genetic variants have been identified increasingly in ticks and vertebrates using molecular techniques, especially in wildlife [7, 17, 29, 30]. In this study, a potential novel Anaplasma species was identified in goat. This isolate was genetically distinct from other known Anaplasma species based on 16S rRNA and groEL genes and is closely related to an unclassified Anaplasma species, which has been identified in H. asiaticum ticks from Xinjiang, northwestern China [27]. The global health burden and risk of anaplasmosis on human and animals seems to be underestimated [31], we may assume that additional novel Anaplasma species remain undiscovered and contribute to human and/or animal diseases. Further investigation is needed with respect to the effects of anaplasmoses on both human and animals.
Conclusions
A molecular survey of A. phagocytophilum was conducted in ruminants from ten provinces in central and southeastern China. Anaplasma phagocytophilum was detected in 106 (25.2%) sheep and goats. Two 16S rRNA variants and a new ecotype (V) of A. phagocytophilum were identified and endemic in small ruminants in China. Moreover, a potential novel Anaplasma species was detected in goat in the present study. Our findings might provide valuable information for the control and management of anaplasmosis in China.
Abbreviations
- DNA:
-
Deoxyribonucleic acid
- EDTA:
-
Ethylene diamine tetraacetic acid
- groEL :
-
heat-shock operon
- HGA:
-
Human granulocytic anaplasmosis
- PCR:
-
Polymerase chain reaction
- UV:
-
Ultraviolet.
References
Chen SM, Dumler JS, Bakken JS, Walker DH. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J Clin Microbiol. 1994;32(3):589–95.
Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, et al. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001;51(Pt 6):2145–65.
Jahfari S, Coipan EC, Fonville M, van Leeuwen AD, Hengeveld P, Heylen D, et al. Circulation of four Anaplasma phagocytophilum ecotypes in Europe. Parasit Vectors. 2014;7:365.
Zhang L, Liu Y, Ni D, Li Q, Yu Y, Yu XJ, et al. Nosocomial transmission of human granulocytic anaplasmosis in China. JAMA. 2008;300(19):2263–70.
Fang LQ, Liu K, Li XL, Liang S, Yang Y, Yao HW, et al. Emerging tick-borne infections in mainland China: an increasing public health threat. Lancet Infect Dis. 2015;15(12):1467–79.
Derdakova M, Stefancikova A, Spitalska E, Taragelova V, Kostalova T, Hrklova G, et al. Emergence and genetic variability of Anaplasma species in small ruminants and ticks from Central Europe. Vet Microbiol. 2011;153(3–4):293–8.
Barakova I, Derdakova M, Carpi G, Rosso F, Collini M, Tagliapietra V, et al. Genetic and ecologic variability among Anaplasma phagocytophilum strains, northern Italy. Emerg Infect Dis. 2014;20(6):1082–5.
Morissette E, Massung RF, Foley JE, Alleman AR, Foley P, Barbet AF. Diversity of Anaplasma phagocytophilum strains, USA. Emerg Infect Dis. 2009;15(6):928–31.
Strasek Smrdel K, Tozon N, Duh D, Petrovec M, Avsic Zupanc T. Diversity of groESL sequences of Anaplasma phagocytophilum among dogs in Slovenia. Clin Microbiol Infect. 2009;15 Suppl 2:79–80.
Strasek Smrdel K, von Loewenich FD, Petrovec M, Avsic Zupanc T. Diversity of ankA and msp4 genes of Anaplasma phagocytophilum in Slovenia. Ticks Tick Borne Dis. 2015;6(2):164–6.
Rar VA, Epikhina TI, Yakimenko VV, Malkova MG, Tancev AK, Bondarenko EI, et al. Genetic variability of Anaplasma phagocytophilum in ticks and voles from Ixodes persulcatus/Ixodes trianguliceps sympatric areas from Western Siberia, Russia. Ticks Tick Borne Dis. 2014;5(6):854–63.
Rar VA, Livanova NN, Panov VV, Doroschenko EK, Pukhovskaya NM, Vysochina NP, Ivanov LI. Genetic diversity of Anaplasma and Ehrlichia in the Asian part of Russia. Ticks Tick Borne Dis. 2010;1(1):57–65.
Liu Z, Ma M, Wang Z, Wang J, Peng Y, Li Y, et al. Molecular survey and genetic identification of Anaplasma species in goats from central and southern China. Appl Environ Microbiol. 2012;78(2):464–70.
Yang J, Liu Z, Guan G, Liu Q, Li Y, Chen Z, et al. Prevalence of Anaplasma phagocytophilum in ruminants, rodents and ticks in Gansu, north-western China. J Med Microbiol. 2013;62(Pt 2):254–8.
Zhan L, Cao WC, Jiang JF, Zhang XA, Liu YX, Wu XM, et al. Anaplasma phagocytophilum from rodents and sheep, China. Emerg Infect Dis. 2010;16(5):764.
Zhan L, Cao WC, Jiang JF, Zhang XA, Wu XM, Zhang WY, et al. Anaplasma phagocytophilum in livestock and small rodents. Vet Microbiol. 2010;144(3–4):405–8.
Kawahara M, Rikihisa Y, Lin Q, Isogai E, Tahara K, Itagaki A, et al. Novel genetic variants of Anaplasma phagocytophilum, Anaplasma bovis, Anaplasma centrale, and a novel Ehrlichia sp. in wild deer and ticks on two major islands in Japan. Appl Environ Microbiol. 2006;72(2):1102–9.
Barlough JE, Madigan JE, DeRock E, Bigornia L. Nested polymerase chain reaction for detection of Ehrlichia equi genomic DNA in horses and ticks (Ixodes pacificus). Vet Parasitol. 1996;63(3–4):319–29.
Alberti A, Zobba R, Chessa B, Addis MF, Sparagano O, Pinna Parpaglia ML, et al. Equine and canine Anaplasma phagocytophilum strains isolated on the island of Sardinia (Italy) are phylogenetically related to pathogenic strains from the United States. Appl Environ Microbiol. 2005;71(10):6418–22.
Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–9.
Rar V, Golovljova I. Anaplasma, Ehrlichia, and “Candidatus Neoehrlichia” bacteria: pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infect Genet Evol. 2011;11(8):1842–61.
Zhan L, Chu CY, Zuo SQ, Wu XM, Dumler JS, Jia N, et al. Anaplasma phagocytophilum and Borrelia burgdorferi in rabbits from southeastern China. Vet Parasitol. 2009;162(3–4):354–6.
Chahan B, Jian Z, Xuan X, Sato Y, Kabeya H, Tuchiya K, et al. Serological evidence of infection of Anaplasma and Ehrlichia in domestic animals in Xinjiang Uygur Autonomous Region area, China. Vet Parasitol. 2005;134(3–4):273–8.
Zhang L, Liu H, Xu B, Lu Q, Li L, Chang L, et al. Anaplasma phagocytophilum infection in domestic animals in ten provinces/cities of China. Am J Trop Med Hyg. 2012;87(1):185–9.
Brown WC, Barbet AF. Persistent infections and immunity in ruminants to arthropod-borne bacteria in the family Anaplasmataceae. Annu Rev Anim Biosci. 2016;4:177–97.
Katargina O, Geller J, Alekseev A, Dubinina H, Efremova G, Mishaeva N, et al. Identification of Anaplasma phagocytophilum in tick populations in Estonia, the European part of Russia and Belarus. Clin Microbiol Infect. 2012;18(1):40–6.
Kang YJ, Diao XN, Zhao GY, Chen MH, Xiong Y, Shi M, et al. Extensive diversity of Rickettsiales bacteria in two species of ticks from China and the evolution of the Rickettsiales. BMC Evol Biol. 2014;14:167.
Li H, Zheng YC, Ma L, Jia N, Jiang BG, Jiang RR, et al. Human infection with a novel tick-borne Anaplasma species in China: a surveillance study. Lancet Infect Dis. 2015;15(6):663–70.
Tate CM, Howerth EW, Mead DG, Dugan VG, Luttrell MP, Sahora AI, et al. Anaplasma odocoilei sp. nov. (family Anaplasmataceae) from white-tailed deer (Odocoileus virginianus). Ticks Tick Borne Dis. 2013;4(1–2):110–9.
Ybanez AP, Matsumoto K, Kishimoto T, Inokuma H. Molecular analyses of a potentially novel Anaplasma species closely related to Anaplasma phagocytophilum detected in sika deer (Cervus nippon yesoensis) in Japan. Vet Microbiol. 2012;157(1–2):232–6.
Home C. Notice to readers: final 2013 reports of nationally notifiable infectious diseases. Neurol Today. 2014;63(32):13–6.
Acknowledgements
Not applicable.
Funding
This study was financially supported by the NSFC (31502091, 31402189, and 31471967); ASTIP, FRIP (2014ZL010), CAAS; NBCIS CARS-38; 973 Program (2015CB150300); and the Jiangsu Co-Innovation Center Program for the Prevention and Control of Important Animal Infectious Diseases and Zoonoses, State Key Laboratory of Veterinary Etiological Biology Project.
Availability of data and material
The datasets supporting the conclusions of this article are included within the article and its additional file. Sequences are submitted in the GenBank database under accession numbers KX272641–KX272643 and KX276166–KX276167.
Authors’ contributions
HY and JL designed this study and critically revised the manuscript. JY participated in its design, coordination and revised the manuscript revision. JY, ZL, QN, GG and JL participated in sample collection. JY, QN, RH, YL and GL performed the experiments, data analysis, and drafted the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethical approval
All experiments with animals were conducted in compliance with the Animal Ethics Procedures and Guidelines of the P. R. China, which was approved by the Animal Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
Author information
Authors and Affiliations
Corresponding authors
Additional file
Additional file 1: Table S1.
GroEL gene sequences of A. phagocytophilum isolates analysed in this study. (XLSX 19 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yang, J., Liu, Z., Niu, Q. et al. Anaplasma phagocytophilum in sheep and goats in central and southeastern China. Parasites Vectors 9, 593 (2016). https://doi.org/10.1186/s13071-016-1880-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-016-1880-z