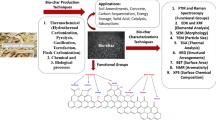
Abstract
The production of biofuels as an efficient source of renewable energy has received considerable attention due to increasing energy demands and regulatory incentives to reduce greenhouse gas emissions. Second-generation biofuel feedstocks, including agricultural crop residues generated on-farm during annual harvests, are abundant, inexpensive, and sustainable. Unlike first-generation feedstocks, which are enriched in easily fermentable carbohydrates, crop residue cell walls are highly resistant to saccharification, fermentation, and valorization. Crop residues contain recalcitrant polysaccharides, including cellulose, hemicelluloses, pectins, and lignin and lignin-carbohydrate complexes. In addition, their cell walls can vary in linkage structure and monosaccharide composition between plant sources. Characterization of total cell wall structure, including high-resolution analyses of saccharide composition, linkage, and complex structures using chromatography-based methods, nuclear magnetic resonance, -omics, and antibody glycome profiling, provides critical insight into the fine chemistry of feedstock cell walls. Furthermore, improving both the catalytic potential of microbial communities that populate biodigester reactors and the efficiency of pre-treatments used in bioethanol production may improve bioconversion rates and yields. Toward this end, knowledge and characterization of carbohydrate-active enzymes (CAZymes) involved in dynamic biomass deconstruction is pivotal. Here we overview the use of common “-omics”-based methods for the study of lignocellulose-metabolizing communities and microorganisms, as well as methods for annotation and discovery of CAZymes, and accurate prediction of CAZyme function. Emerging approaches for analysis of large datasets, including metagenome-assembled genomes, are also discussed. Using complementary glycomic and meta-omic methods to characterize agricultural residues and the microbial communities that digest them provides promising streams of research to maximize value and energy extraction from crop waste streams.
Similar content being viewed by others
Background
Growing international concern over climate change has led to continued interest in generating bioliquids (e.g., ethanol) and biogases (e.g., methane) from viable and sustainable sources of energy. First-generation biofuel crops, such as corn and sugarcane, which contain high amounts of starch and sucrose, respectively, are readily fermented by microorganisms to produce ethanol and biogas in biodigesters [1, 2]. However, their use for biofuel production has socioeconomic consequnces , including the food versus fuel debate, as their dedicated use for fuel directly impacts food prices and competition of land usage [3]. Second-generation biofuel crops do not compete directly with food production and have been well regarded as sustainable sources of fermentable biomass. These feedstocks include inedible woody plants, bioenergy crops (e.g., switchgrass), and agricultural residues.
Crop residues are biomaterials remaining in the field after harvest and consist mainly of straw or stover from grains and oilseeds. Primary sources include rice (Oryza sativa), wheat (Triticum aestivum), corn (Zea mays), barely (Hordeum vulgare), oat (Avena sativa), rye (Secale cereale), canola (Brassica napus), flax (Linum usitatissimum), peanut (Arachis hypogaea), sunflower (Helianthus annuus), sorghum (Sorghum bicolor), soybean (Glycine max), pea (Pisum sativum), and chickpea (Cicer arietinum) [4,5,6,7,8,9,10,11,12]. Historically, crop residues are usually left to decay on field after threshing and were incorporated into soil by plowing and disking or used as livestock feed or bedding [13]. Seasonal burning of agricultural residues is practiced in many countries, resulting in large scale wastage and has been linked to environmental problems, such as emission of airborne particulate matter (PM) pollutants (e.g., PM2.5) and greenhouse gases [14, 15].
Crop residues are readily available and produced in great quantities. Globally, the total residue produced from a collection of 27 common food crops was estimated to be 3.8 billion tonnes per year [16], and the theoretical global energy potential from six major crop residues was estimated to be 65 exajoules per year, equaling 66% of annual worldwide transportation energy consumption in 2006–2008 [7]. However, the high concentration of lignocellulosic biomass, including recalcitrant polysaccharides, such as cellulose, hemicelluloses, pectins, and aromatic polymers (i.e., lignin), has limited their widespread use in biofuel production. Cross-linking of hemicellulose to lignin and hemicellulose–cellulose interactions further contribute to biomass recalcitrance [17]. Moreover, the diversity of monosaccharide composition and non-cellulosic carbohydrate lignin linkages can vary between crop residues [18], affecting their valorization as high-value products, including ethanol and methane.
Carbohydrate-active enzymes (CAZymes) are commonly used in biofuels to convert recalcitrant polysaccharides into fermentable carbohydrates. In bioethanol production, CAZymes are added to biomass prior to or simultaneously with fermentation, or expressed from an engineered organism for consolidated bioprocessing [19]; whereas biogas production uses the native production of CAZymes from anaerobic microorganisms within a biomass biodigester [2]. To date, numerous CAZyme classes and families have been discovered that target cellulose and other plant cell wall polysaccharide linkages in biofuel feedstocks [20]. Enabling technologies and software to sequence genomes/metagenomes and annotate/predict novel CAZymes have resulted in extensive literature describing new CAZymes and microorganisms for biorefinery applications.
Two areas that are pivotal for valorization of agricultural residues as viable feedstocks are: 1) to elucidate the carbohydrate composition and linkages within the plant cell wall material, and 2) to optimize enzyme, microbe, or microbial community treatments to maximize release of fermentable carbohydrates. This review will focus on recent analyses of common crop residue cell wall structures, current glycomic methods used for cell wall analysis, and in silico assessment of CAZyme function, or lack thereof, encoded within microbial communities to inform more efficient polysaccharide saccharification.
Crop cell wall polysaccharides
The cell wall material of agricultural residues is comprised predominantly of cellulosic, hemicellulosic, and pectic polysaccharides, of which cellulose predominates. Cellulose is a linear chain of 4-linked β-D-glucopyranoses existing abundantly in the form of hydrogen-bonded, cable-like microfibrils that contain a heterogeneous mixture of crystalline and amorphous regions with a diameter ranging from 3 to 20 nm depending on cell wall type [21]. Non-cellulosic polysaccharides demonstrate great diversity in monosaccharide composition and linkage (Fig. 1). Hemicelluloses are a group of plant polysaccharides consisting mostly of 4-linked neutral sugar backbone, with or without side chains or substituent groups (e.g., methyl group, acetyl group, and ferulic acids). This includes mainly xyloglucan, xylan, and heteroxylans (e.g., arabinoxylan (AX), 4-O-methyl glucuronoxylan (GX), glucuronoarabinoxylan (GAX)), mannans, and heteromannans (e.g., glucomannan (GlM), galactomannan (GaM), and galactoglucomannan (GGM)), and mixed-linkage glucans in higher plants [21, 22]. Callose is a linear 3-linked β-D-glucan, and although its classification of a hemicellulose is debated, it is important in higher plant cell development and responses to environmental cues [21, 23]. Pectins are a group of galacturonic acid-rich polysaccharides, including homogalacturonan (HG) and rhamnogalacturonans (RG-I and RG-II). HG has a 4-linked galacturonic acid backbone that can be 6-O-methyl-esterified and O-acetylated [21]. RG-I consists of a backbone of alternating galacturonic acids and rhamnoses and side chains of arabinan, galactan, and arabinogalactans, while RG-II is composed of a homogalacturonan backbone decorated with highly complex side chain structures built with more than 20 types of glycosidic linkages from 13 different monosaccharides [21, 24]. Aside from the wide variety of monosaccharide and linkage composition between polysaccharides, the cell wall becomes increasingly complex when considering inter- and intra-chain interactions between polysaccharides. Cellulose microfibrils commonly interact with pectin and hemicelluloses (xylans, mannans, and xyloglucan) through hydrogen bonding [25]. Pectins are also known to gel and interact with one another in the presence of calcium and boron [26]; as well, cross-linking within arabinan chains in pectins [27] and AX chains [28] by feruloyl residues has been well noted. Structural variation is complex and has been extensively studied and reviewed [21, 29]. Importantly, variations in the fine chemistry of these networks exist between plant species and at different developmental stages [30].
Crop cell wall polysaccharide variation
Monocot (cereal crops, such as corn, wheat, and barley) and dicot plants (legumes, oilseeds, and soybeans) have similar cellulose content in primary and secondary cell walls, but differ greatly in the abundance and chemistry of hemicelluloses [31,32,33,34]. Typically, monocots contain much more heteroxylans than dicots in both the primary (20–40 vs. 5%) and secondary cell wall (40–50 vs. 20–30%) [31, 35,36,37,38,39]. Heteroxylans can vary greatly in their substitution patterns, effecting interactions with cellulose and lignin, and in turn, biomass recalcitrance [17, 40]. Dicots generally contain more GX, whereas monocot heteroxylans contain arabinose sidechains (AX and GAX) [18]. This difference can be observed between common agricultural crops, including canola, a dicot [41], and cereals [42]. Mixed-linkage glucans are absent in most dicots, but represent 10–30% of total cell wall content in monocots [31, 39]. This is in contrast to xyloglucan (20–35 vs. 5%) and pectins (20–25 vs. 1–5%), which are more prevalent in primary cell walls of dicots rather than monocots [31, 36, 39]. Although large differences in hemicellulose content and composition exist between monocot and dicot plants, variation can also be seen within a single group. For example, monocot heteroxylans can differ in concentration, presence of GAX or AX, and xylan substitution level or arabinose:xylose ratio [42,43,44,45]. Furthermore, variation can be seen at the species level as xyloglucan sidechains were shown to differ between canola species B. napus and B. campestris [41], between plant anatomy (e.g., root vs. root hairs; sugarcane bagasse vs. straw) [46, 47], and between developmental stages in rice [48].
Plant cell wall polysaccharides are not the only structural differences observed; cross-linking between structural carbohydrates by lignin are also diverse. Lignin is a hydrophobic, polyphenolic biopolymer consisting mainly of three phenylpropanoid monomers with varying degrees of methoxylation, including p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units [49]. Lignin increases cell wall recalcitrance by forming complex interactions with plant cell wall hemicelluloses, including heteroxylans in monocots and heteromannans in dicots [17] (Fig. 1). Lignin in monocot crops contains substantially more ferulic and ρ-coumaric acid than in dicots [31]. These components form covalent linkages with arabinose sidechains on GAX and AX; however, lignin can also be conjugated to the backbone of GGM [17, 31].
Notably, the structural diversity of plant cell wall polysaccharides and lignin polymers that exists in nature can be further augmented by common pre-treatments that cause chemical modification of cell wall polysaccharides [34]. Thus, a comprehensive understanding of plant cell wall chemistry is helpful throughout the treatment process.
Cell wall analysis techniques
Glycomic analysis of plant cell walls has seen a recent resurgence in part due to the demand for using plant biomass for biofuels [22]. These methods have improved and proven useful in elucidating the structure of native crop plant cell wall polysaccharides [50], modifications resulting from pre-treatments, and biodigester waste residues [51,52,53]. Glycomic analysis of lignocellulose can range from composition (e.g., total sugar, total lignin, monosaccharide composition, and lignin monomer composition) to detailed structural features (e.g., glycosidic linkage composition and sequences; lignin–carbohydrate interaction) with the use of advanced analytical instruments and techniques described below and summarized in Fig. 2.
Analytical methods for total cell wall analysis. a UV/Vis spectrophotometer colorimetric assays. AX*: total arabinoxylan can be determined through commercially available kit; b HPAEC-PAD; c GC–MS/FID; d LC–ESI–MS/MS; e NMR; and f Immunological methods, such as Glycome profiling and MAPP. Corn GAX was used as a model polysaccharide to demonstrate representative structural information that could be inferred by each method [28]
UV–Vis spectrophotometer
Colorimetric assays (Fig. 2a) can be performed using a simple UV–Vis spectrophotometer for quantification of neutral carbohydrates [54], uronic acids [55, 56], lignins [57], and substituents groups (e.g., ferulate and acetate) [58,59,60] of whole plant cell walls prepared from agricultural residues. A broad range of enzymatic–colorimetric assay kits are commercially available (e.g., Megazyme, Sigma-Aldrich) for the analysis of starch and non-starch polysaccharides, such as arabinan, AX, mixed-linkage glucan, GlM, and GaM in lignocellulosic biomass of agricultural residue.
High-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD)
HPAEC-PAD (Fig. 2b) is convenient for the identification of liberated neutral monosaccharides and uronic acids from plant residues [61]. Neutral sugars from non-cellulosic components of agricultural residue can be readily hydrolyzed by trifluoroacetic acid (TFA) into alditol acetates for analysis (e.g., 2 M, 120 °C, 2 h) [22, 62]; however, sulfuric acid is normally used for the complete hydrolysis of recalcitrant crystalline cellulose in agricultural residue [22]. Methanolysis combined with TFA hydrolysis is best suited for water-soluble uronic acid-containing polysaccharides [63, 64]. Complementary to HPAEC-PAD, reverse-phase high-performance liquid chromatography coupled to ultraviolet detection (RP-HPLC–UV) with various pre- or post-column derivatization approaches (e.g., 1-phenyl-3-methyl-5-pyrazolone) are available for monosaccharide analysis [65, 66]. A benefit of HPAEC-PAD is that it does not require derivatization; it is more commonly used than the RP-HPLC–UV method for monosaccharide analysis of plant residues. In addition to monosaccharide analysis, HPAEC-PAD is an important method for detecting and quantifying oligosaccharides and evaluating the purity of purified oligosaccharide samples [67, 68].
Gas chromatography–mass spectrometry/flame ionization detection (GC–MS/FID)
GC–MS/FID (Fig. 2c) is an essential tool for the monosaccharide analysis of agriculture residues. Over the past several decades, many derivatization methods have been developed for GC–MS/FID analysis of monosaccharides [69]. Among them, the alditol acetate (AA) derivatization method is the most common [70]. Notably, a GC–MS procedure has been recently developed for comprehensive monosaccharide analysis of insoluble lignocelluloses resistant to acid hydrolysis based upon alditol acetate derivatization [71]. Glycosidic linkage analysis, normally referred to as “methylation analysis,” is a fundamental technique for structural characterization of plant cell wall polysaccharides based on GC–MS/FID analysis of the partially methylated alditol acetate (PMAA) derivatives prepared by permethylation, hydrolysis, reduction, and peracetylation of whole cell wall and fractions [22, 70, 72, 73]. Uronic acids in plant residues are converted to their corresponding 6,6-dideuterio neutral sugars before methylation analysis [74, 75]. Deuteriomethylation or ethylation is used for localizing the naturally existing O-methyl group during linkage analysis of cell wall polysaccharides (e.g., 4-O-methylglucuronic acids of GX) [76,77,78]. The relative composition of plant polysaccharides can be estimated from the results of linkage composition by assigning glycosidic linkages to corresponding polysaccharide structures followed by summing up all the values grouped to each structure [22].
Liquid chromatography electrospray ionization tandem mass spectrometry (LC–ESI–MS/MS)
LC–ESI–MS/MS (Fig. 2d) is most commonly used for determining the molecular mass and linkage sequence of oligosaccharides generated by partial depolymerization of cell wall polysaccharides through enzymatic and/or chemical means (e.g., weak acid hydrolysis, methanolysis, acetolysis, alkaline degradation, and β-elimination) [79]. Oligosaccharides are usually purified using graphitized carbon solid-phase extraction before structural characterization by LC–ESI–MS/MS [67]. NMR and other MS techniques (e.g., MALDI-tof–MS) are complimentary to LC–ESI–MS/MS for structural analysis of oligosaccharides released enzymatically or chemically from plant residues [67, 68, 80]. Recently, there has been interest in the development of LC–MS-based methods for glyosidic linkage analysis [81,82,83,84,85], and LC–ESI–MS/MS methods have been developed for fast monosaccharide analysis with high sensitivity [86,87,88]. These novel methylation-LC–MS analyses are fast and sensitive and can be used to complement current GC-based linkage analyses.
Nuclear magnetic resonance (NMR)
Advanced structural features (e.g., anomeric configuration, ring forms, substituents, glyosidic linkage composition, and sequence) of polysaccharides isolated from agricultural residues can be obtained by a series of one-dimensional (1D), two-dimensional (2D) (e.g., COSY, TOCSY, HSQC, HMBC, NOESY, and ROESY), and three-dimensional (e.g., TOCSY-HSQC) solution-state NMR experiments (Fig. 2e; [89, 90]). A recently developed method involving permethylation followed by 2D 1H-13C HSQC solution-state NMR analysis can be used for polysaccharide profiling of whole cell wall [91]. A novel method for collecting 2D 1H-13C HSQC NMR spectra from non-derivatized ball-milled whole cell wall dissolved in deuterated reagents (e.g., DMSO-d6/pyridine-d5) has been increasingly popular for lignocellulose characterization [49, 92,93,94,95]. Impressive progress has been made within the past decade in solid-state NMR analysis by the production of uniformly isotope-labeled plant and fungi cell wall samples by feeding 13CO2 or media containing 13C-glucose and 15N-salts, and by the introduction of ultrahigh-field (e.g., 900 MHz) NMR spectrometers [40, 96, 97]. For instance, recent high-resolution multi-dimensional magic-angle spinning solid-state NMR evidence indicated that cellulose, hemicelluloses, and pectins could be associated non-covalently with the sub-nanometer scale to form an integrated network in plant primary cell walls [97]. A series of high-resolution solid-state 2D 13C-13C correlation NMR methods specifically designed for enhancing the detection of lignin aromatic signals were successfully used for the structural characterization of lignin–carbohydrate interface of plant secondary cell walls (e.g., mature stems of rice, maize, and switchgrass) [98].
Glycome profiling/microarray polymer profiling (MAPP)
Large collections (more than 200 worldwide) of cell wall glycan-directed monoclonal antibodies (mAbs) with known glycan epitope-binding specificities have allowed for the development of immunological methods for screening plant cell wall samples, termed glycome profiling (Fig. 2f; [99]). This analysis is conducted on fractionated plant cell walls using increasingly harsh chemicals, followed by an ELISA of the fractions in a 96-well plate; the results are commonly presented as a heat map [99]. Alternatively, a microarray polymer profiling (MAPP) procedure involving the integration of cell wall sequential fractionation with the generation of microarrays probed with glycan-binding mAbs or carbohydrate-binding modules (CBMs) has been developed [100]. Both immunological procedures have proved to be very useful for high-throughput screening of whole cell wall polysaccharides and their degradation products during and after bioconversion, and can be used in combination with other polysaccharide screening techniques, such as Fourier transform infrared spectroscopy-attenuated total reflectance [101,102,103,104,105].
CAZymes in the production of biofuels
CAZymes are classified based upon the catalytic mechanism by which they act, including glycoside hydrolases (GH) [106, 107], polysaccharide lyases (PL) [108], carbohydrate esterases (CE) [108], and auxiliary activities (AA) [109] (Fig. 3a). Each of these classes are further divided into sequence-related families.
CAZyme depolymerization mechanisms and specificities. a Simplified reaction schematics are shown of a glycoside hydrolase (GH), polysaccharide lyase (PL), carbohydrate esterases (CEs) acetyl (top) and methyl (bottom), and the auxiliary activities (AA) of LPMOs active on C1 and C4. b CAZyme-targeted bonds of plant cell wall polysaccharides homogalacturonan (HG), cellulose, and corn GAX [28]) are shown, with example CAZy family and enzyme class (EC) numbers as indicated
GHs hydrolyze glycosidic bonds between carbohydrates or a carbohydrate and aglycone moiety, such as lipids or proteins [106, 107]. For most GH-mediated hydrolysis, two residues are critical for this enzymatic mechanism, a proton donor and a nucleophile/base, and results in a mechanism that either retains or inverts the anomeric configuration [106, 110]. With such diverse substrate potential existing in nature [111], it is unsurprising that GHs have been found to be active on carbohydrate polymers ranging from homopolymers, such as starch [112] and cellulose (Fig. 3b) [113], to highly branched and chemically heterogeneous substrates, such as pectins [24, 114]. At the time of publication, GHs have been classified into 168 sequence-based families in the CAZy database [115].
PLs cleave polysaccharide chains with a β-elimination reaction, resulting in a terminal hexenuronic acid [108, 110]. PLs are typically involved in the cleavage of acidic substrates, such as pectins (e.g., HG; (Fig. 3B)), chondroitin, xanthan, and alginate [116]. At the time of publication, 40 different PL families have been assigned within the CAZy database [115].
CE families are currently classified into 18 different families [115]. These enzymes catalyze the de-O- or de-N-acetylation of esterified sugars through a variety of mechanisms, whereby the sugar can either act as the acid (e.g., pectin methyl esters) or the alcohol (e.g., acetylated xylan) (Fig. 3b; [110]). Removal of carbohydrate esters increases the access of GHs and PLs to their substrates, and therefore is an important event in the catabolism of chemically complex polysaccharides.
AAs are the most recently described CAZyme class and deploy a redox reaction to fragment structural polysaccharide and lignin substrates [109]. AAs are currently divided into 16 families, encompassing 9 families of ligninolytic enzymes, and 6 families of lytic polysaccharide monooxygenases (LPMOs), and while first [20] discovered to target chitin [117], LPMOs have demonstrated activity on common plant cell wall polysaccharides including cellulose. (Fig. 3b). Many AA enzymes are metalloenzymes, requiring copper to catalyze the digestion of lignocellulosic biomass [118, 119].
Cellulose-active CAZymes
Cellulose is the most homogeneous and abundant source of glucose in agricultural biomass. Despite its simple β-1,4-linked glucose repeating structure, the crystalline higher-order structure of cellulose limits the access to cellulose-degrading CAZymes [120]. However, synergistic effects are observed when multiple enzymes are used in combination on intact cellulose, which can help overcome poor enzyme efficacy [121, 122]. Combined strategies, involving several different exo- and endo-acting GHs are used for efficient saccharification [123,124,125,126]. Endo-β-1,4-glucanases (enzyme class (EC) 3.2.1.4) cleave internal bonds within the cellulose chains and represent most enzymes used for the hydrolysis of glucosidic linkages in cellulose, while cellobiosidases (EC 3.2.1.91) processively release disaccharides from cellulose chains. Cellobiose and cellooligosaccharides released are further depolymerized by endo-β-glucosidases (EC 3.2.1.21), cellodextrinases (EC 3.2.1.74), and cellobiose phosphorylases (2.4.1.20). Cellodextrinases are preferentially active on longer substrates and hydrolyze terminal, non-reducing β-d-glucosyl residues from cellulose in a step-wise fashion [127].
GH5, GH6, GH7, GH9, GH12, and GH45 CAZy families contain most cellulose-active hydrolases [115, 128, 129]. GH5 is one of the largest polyspecific GH families in the CAZy database. Once known as “cellulase family A,” it is now known to contain a variety of catalytic specificities, including endo-glucanase, as well as many others, including endo-mannosidase (EC 3.2.1.78), endo-xylanase (EC 3.2.1.8), and endo-β-1,6-glucosidase (EC 3.2.1.75). As such, the GH5 family has been further subdivided into sequence-related subfamilies to better classify conserved specificities [130] (Fig. 4a). The GH6 family consists solely of endo-glucanases and cellobiohydrolases, which also compose most of the GH7 family. GH9 is the second largest family of cellulase enzymes, comprised primarily of endo-glucanases. Endo-glucanases are found in the GH12 family, among xyloglucan endo-transglycosidase and xyloglucan endo-hydrolase activities. Finally, GH45 family members function as endo-glucanases; however, some are specific to xyloglucan.
Polyspecific CAZy families GH5 and GH43. Phylogenetic trees were built using SACCHARIS [195] with characterized sequences for a GH5 and b GH43 CAZy families. Annotations were generated using ITOL [228]. Enzyme activities, for example, subfamilies, are labeled with the corresponding EC numbers, and targeted substrates are illustrated by cartoons following the Symbol Nomenclature for Glycans [227]
Weak endo-glucanase activity was seen in the GH61 and CBM33 family. However, both these families are now understood to be LPMOs, which target cellulose through oxidative cleavage. GH61 has been reclassified as AA9 [131], while CBM33 has been reclassified as AA10 and is known to possess enzymes active on cellulose or chitin [117].
Hemicellulose- and pectin-active CAZymes
Due to the abundance of xylan in plant cell walls, there has been a concerted effort to understand xylan and heteroxylan digestion by endo-β-1,4-xylanases (EC 3.2.1.8), β-1,4-xylosidases (EC 3.2.1.37), arabinan endo-α-1,5-l-arabinanases (EC 3.2.1.99), and non-reducing end α-l-arabinofuranosidases (EC 3.2.1.55).
GH10 and GH11 predominantly contain endo-β-1,4-xylanases, and enzymes from these families work synergistically to break down xylan and heteroxylan. GH11s are active on xylans at least seven sugars in length, while GH10s are better suited to the hydrolysis of xylosyl linkages close to arabinosyl-substitutions [132]. As well, in highly substituted wheat bran AX, GH10 xylanases are able to accommodate arabinose decorated xylose residues, whereas GH11 xylanases do not [133].
AX is a large component of monocot hemicellulosic polysaccharides and thus a common substrate for arabinofuranosidases and arabinanases. GH43 is a polyspecific family divided into many subfamilies [134] (Fig. 4b) and contains many α-l-arabinofuranosidases and α-l-arabinanases active on AX. Arabinofuranosidases have been classified based on substrate determinants [132]:: (1) type A, active on pNP-α-l-arabinofuranosides and short arabinooligosaccharides; (2) type B, active on short oligosaccharides and longer polysaccharides, such as arabinan and AX; and (3) AX arabinofuranohydrolases. Recent studies have shown that rumen fungi are adept at producing GH43 enzymes for the breakdown of complex hemicelluloses, and these enzymes may represent the most abundant fungal glycoside hydrolases for these reactions [135].
CE enzymes (e.g., acetyl xylan esterase EC 3.1.1.72, feruloyl esterase EC 3.1.1.73, CE families 1 through 7) can facilitate accessibility of hydrolytic enzymes to their substrates, as large modifications, substitutions, and cross-linking of carbohydrate residues impede enzymatic catalysis. For example, corn bran is highly recalcitrant to enzymatic digestion [136, 137], likely due to ferulate cross-links within AX [138], but the inclusion of acetyl xylan esterases (CE1) and feruloyl esterases (CE1), alongside xylanases (GH10), xylosidases (GH3), and arabinofuranosidases (GH43, GH51) significantly increased the release of total monomeric xylose [28]. The cooperation between the different enzyme activities of CEs and GHs may be necessary for the complete hydrolysis of heavily modified hemicellulosic and pectic polysaccharides. Interestingly, there is some recent evidence to suggest that LPMOs are also active on xylans and xyloglucans and contribute to the large array of catalytic strategies evolved to dismantle these complex substrates [139].
Modifying plant genetics to reduce recalcitrant residues
Glycosyltransferases (GTs) are responsible for the synthesis of structural polysaccharides, storage polysaccharides, and other complex glycans [140]. The formation of glycosidic bonds involves the transfer of a carbohydrate moiety from sugar donors to acceptor molecules [110], and cascading glycosylation by downstream GTs results in increasingly complex carbohydrates. For example, biosynthesis of plant pectic polysaccharides requires hundreds of glycosyltransferases to produce the extensive variety of glycosidic linkages and adducts [141]. Genetic manipulation of these biological processes can reduce the number of recalcitrant residues in the plant cell wall [17], namely cellulosic [142] and hemicellulosic [18] biomass. Initial attempts have been made as an alternative to enzymatic treatment, such as the downregulation of GT8 family pectin biosynthetic genes in switchgrass which leads to decreased lignocellulose and pectin cross-linking, thereby reducing the recalcitrance of biomass [143, 144].
Strategies for CAZyme-catalyzed digestion of lignocellulosic biomass
Interactions between cellulose, hemicellulose, pectin, and lignin leads to a complex network that is highly recalcitrant to enzymatic deconstruction. Studies have begun to look at the hydrolysis of these interactions by enzymes, such as AA family LPMOs [131]. Additionally, AA lignin-modifying enzyme families may have a role; laccases, manganese peroxidases, and lignin peroxidases all potentially contribute to the modification of cross-links and subsequent delignification, exposing the underlying polysaccharides for further modification by GH and CE enzymes [145]. Along with feruloyl esterase, CE15 glucuronyl esterases also contribute to the disassembly of lignin–carbohydrate complexes via the cleavage of ester bonds between alcohol and 4-O-methyl-glucuronoyl moieties of lignin and xylan, respectively (Fig. 3b) [146]. Degradation of lignocellulosic biomass has improved using cellulolytic enzyme cocktails [147] and combining lignin-active enzymes with polysaccharide-specific enzymes may be the best strategy for the optimal digestion of complex lignocellulose [148]. Tailoring the mixture to the agricultural residues of interest, and the specific polysaccharides and glycosidic linkages, may be optimal for converting these biological residues into valuable products.
Lignocellulose deconstruction in bioethanol production employs extensive heat treatments to expose biomass for efficient enzymatic attack, often at temperatures above 55 °C [127]. Thus, enzymes are often sourced from thermophilic microbes as they are the most likely to retain properties beneficial for bioprocessing. For example, a GH5 endo-glucanase from Talaromyces emersonii was found to have optimal activity at pH 4.8 and 80 °C, but retains activity for 15 min at temperatures up to 100 °C [149]. Furthermore, non-enzymatic processes that decrease the crystallinity of cellulose typically involve low pH, organic solvents, chemical and oxidative reagents, and detergents [127]. Some enzymes, such as two thermostable cellulases of Melanocarpus albomyces, are more active on crystalline cellulose than amorphous cellulose [150]. These conditions and enzymatic properties need to be taken into consideration when selecting enzymes for the treatment of biomass residues.
-Omic and bioinformatic approaches to elucidate CAZyme function
Extensive research has been invested toward identifying CAZymes, microorganisms, and microbial communities that are capable of saccharifying lignocellulose to reduce the cost and increase the yield of biofuel production. Commonly, organisms selected for fermentation (e.g., Saccharomyces cerevisiae) lack the ability to metabolize lignocellulose [151]. Fungi and bacteria, including the well-studied T. reesei and Clostridium spp., are used to produce lignocellulosic CAZymes [152, 153], as they can secrete large quantities of endogenous cellulolytic CAZymes (i.e., endo-glucanases, exo-glucanases, glucosidases [152], and LPMOs [154]). These CAZymes have greatly increased the efficiency of ethanol production, but the cost of producing and purifying enzymes can make the process economically untenable [19]. To provide affordable solutions for optimized lignocellulose degradation, it is common to bioprospect microbial ecosystems of biodigester systems involved in plant biomass saccharification to identify lignocellulose-degrading microorganisms and their endogenous CAZymes. Promising microbes and/or CAZyme targets have been discovered in crop soil [155], compost [156], wastewater sludge [157], and herbivorous animal microbiomes [158, 159]. Significantly, the anaerobic environment of the ruminant digestive tract and the termite hindgut has led to the discovery of novel species and microorganisms, including the obligate anaerobic fungi phylum Neocallimastigomycota in cattle rumen [160] and lignocellulosic microorganisms found in and cultivated by termites [159, 161]. Microbial analysis of anaerobic environments is of particular interest to the bioethanol and biogas industries due to the parallels that exist between these environments. Moreover, biogas biodigesters are enriched with lignocellulose-degrading organisms as they are optimized for biomass metabolism. Microorganisms and/or CAZymes identified within biodigesters can be used as supplements to further increase the valorization of biodigester feedstocks (Fig. 5). Crop residues, including corn stover [162], barley straw [163], rice straw [164], and wheat straw [165], are commonly used as biodigester feed stocks. However, microbial community composition can vary greatly between systems depending on pH, temperature, and feed substrates [2, 166].
Combinatorial assessment of cell wall structure and investigation of microbial CAZyme function. The integration of analytical methods can be implemented to provide a comprehensive experimental workflow to improve bioconversion of agriculture residues. Crop residues can be studied prior to or after processing using total cell wall analysis. Information on the structure of waste residues can be compared to starting material to determine recalcitrant structures that are limiting the efficiency of bioconversion. The microbial ecosystem of biodigesters can be studied using -omics techniques, such as metagenomics, metatranscriptomics, and metaproteomics, to define the structure and function at the community, microbe, and CAZyme levels. Information gathered using these techniques can inform optimized conditions or identify lacking catalytic functions in the reaction cascade. Microbial communities, microorganisms, and CAZymes can be deployed back into production processes to augment inefficent or absent catalytic reactions and improve biofuel production. Surface representation of enzyme structure (white) was generated using PyMOL [229] (PDB ID: 2CKR), with cellotetrose ligand illustrated in sticks (blue)
Lignocellulose-metabolizing microorganisms can exhibit varied growth conditions depending on their taxonomy and the environment they were isolated from [167], making the cultivation of organisms and discovery of novel CAZymes encoded within their genomes difficult. However, with the recent advances in -omics technologies and decreases in associated costs, the study of complex communities has become more accessible. Metagenomics [168,169,170], metatranscriptomics [163, 166, 171], and metaproteomics [162, 172, 173] have demonstrated the utility of -omics technologies for the discovery of lignocellulose microorganisms and CAZymes. When combined with reference genomes or metagenomes, metatranscriptomics and metaproteomics allow for accurate functional assignment of genes and proteins, respectively [174]. Recent advances in metagenomic sequencing and contig binning have ushered in a new era of metagenomic-assembled genomes, allowing for increased understanding of microbial function within and between microbial ecosystems [175, 176]. For example, a large-scale metagenomic study demonstrated the diversity of species between anaerobic digesters and the importance of generating metagenomic assembled genomes to study and standardize a core and accessory digester microbiome, allowing for efficient optimization of biogas production [177]. Metagenomics and associated software for annotation and functional prediction have also aided in the assembly of eukaryotic genomes in complex environments, which overcomes the historical challenge of sequencing eukaryotic genomes [178]. Genomic and metagenomic databases have rapidly expanded and will continue to do so as the affordability and accessibility of second- and third-generation sequencing technologies increase. Unfortunately, subsequent biochemical characterization of annotated genes has been unable to keep pace with sequencing data. Therefore, accurate and automated annotation of these sequences has become a priority for streamlining CAZyme discovery.
CAZyme annotation and curation
Wide-ranging guidelines have been proposed for unifying how metagenomic studies are performed, covering aspects from sample collection and metagenomic binning [179, 180] to standards for metagenomically generated genomes [175]. Additionally, there are renowned software pipelines for the prediction and annotation of prokaryotic and eukaryotic genes, including PROKKA [181], RAST [182], MAKER2 [183], AUGUSTUS [184], and the NCBI online annotation platforms [185]. Annotation platforms, such as COG [186], SEED [187], Pfam [188], and KEGG [189], have also been instrumental for predicting gene function. However, these platforms are not specialized for CAZyme annotation, nor are they designed to differentiate between the rapidly expanding lists of CAZyme families.
The CAZy database was launched in 1999, and is the single source for CAZyme curation [20]. In addition, it provides links to relevant publications and other online resources, such as CAZypedia [190] and the polysaccharide utilization loci (PUL) database PULDB [191]. These resources have enabled other external platforms to assist with CAZyme discovery and characterization. For example, the CAZyme annotation tool dbCAN [192] provides hidden Markov models (HMMs) generated from the CAZy database to facilitate user sequence annotation. dbCAN identifies sequence boundaries to improve prediction accuracy, creating profile HMMs based on homologous sequence alignments. Alternatively, the CAZyme analysis toolkit [193], currently unmaintained, implements Pfam-defined profile HMMs which were recently shown to identify > 98% of GHs in the CAZy database [194]. These profile HMMs provide valuable protein domain prediction, especially helpful in determining boundaries in multi-modular CAZymes and/or attached CBM modules [195], and are currently used by an expanding list of pipelines and software tools [195,196,197]. However, it should be noted that due to differing thresholds between profile HMMs, there may be discrepancies between Pfam and dbCAN annotations when compared to those of CAZy [20].
The addition of subfamily designations to large, polyspecific families in the CAZy database and the subsequent profile HMMs generated by dbCAN have greatly improved functional prediction of novel sequences for CAZy families GH5 [130], GH13 [198], GH16 [199], GH30 [200], and GH43 [134]. However, there are still inherent limitations with family- and subfamily-based classifications. While members with CAZy families possess the same fold and catalytic mechanisms, assignment of a sequence to a CAZy family is not necessarily definitive of enzyme specificity. Functional differences between members of the same subfamily and polyspecific families without subfamily delineations convolute prediction of CAZyme activity. As well, sequence-based CAZyme prediction is hampered by the low abundance of characterized sequences in the database and variability in substrate libraries used to biochemically characterized enzymes. In this regard, a standardized approach using similar substrates and kinetic parameters to report rate would be beneficial. Fortunately, there is a growing list of novel software packages designed to aid in the annotation (PULpy [201], DRAM [202], and dbCAN-PUL [203]), curation (dbCAN-PUL [203]) and high-resolution phylogeny (SACCHARIS [195], CUPP [196]) of uncharacterized CAZymes.
Both PULpy and DRAM software packages use profile HMMs sourced from both dbCAN and Pfam to identify CAZymes. PULpy focuses heavily on identifying polysaccharide utilization loci (PULs) within metagenomes, demonstrated in ruminants [169], and DRAM extrapolates CAZyme annotation to predict carbohydrate utilization of identified taxonomic units. Recently, dbCAN-PUL was developed for the curation of PULs by substrate, taxonomy, and characterization method. The repository can also be downloaded and used as a database to BLASTX against novel CAZymes. Alternatively, SACCHARIS is a pipeline that streamlines identification and phylogenetic analysis of CAZyme sequences. Sequences collected from the CAZy database, as well as user input sequences, are trimmed to the predicted catalytic domain using dbCAN, aligned [204], and a best-fit Newick tree is generated [205,206,207] (Fig. 4). SACCHARIS is a real-time software which enables the functional prediction of CAZymes based upon tree topologies generated using the current state of knowledge [80, 208, 209]. The Conserved Unique Peptide Patterns (CUPP) downloadable software uses peptide pattern recognition to find conserved peptide motifs within CAZyme families to develop strict CUPP groups or subfamilies, and a recent web server allows for annotation of user sequences [210]. CUPP has been used to elucidate sequence function in pectin and alginate lyase families [211, 212], as well as using fungal CAZyme secretomes to predict fungal phylogenies [213]. Together with -omics-based technologies, CAZyme prediction tools will aid in the interpretation of sequence datasets at the microbe, community, and gene level. Ultimately, this interpretation is necessary to inform CAZyme discovery and characterization, which can be used to improve biofuel production (Fig. 5).
Glycomic and multi-omic integration
Methods to resolve the fine chemistry of biofuel feedstocks and to optimize the valorization of feedstocks through discovery of microorganisms and CAZymes have led to significant advances in biofuel production. Combining these approaches will help unlock further solutions for optimizing the synthesis and saccharification of recalcitrant biomass. Comparative genomics of plant cell wall biosynthetic loci is a complementary approach to glycomics to help illuminate the structural diversity of cell walls that exists between species [30]. Plants employ a wide variety of CAZymes to synthesize, remodel, and saccharify plant cell walls during growth and development [214, 215], and -omics can be used to identify functional orthology between cell wall biosynthetic genes [216]. A multi-tiered approach that includes plant cell wall profiling and CAZyme gene mining has been proposed to better understand cell wall variability between plant species [215]. Recently, CAZyme phylogeny and characterization have been supplemented with analytical methods to investigate acetyl xylan synthesis [217], and variable expression of xylan synthesis glycotransferases between species [218]. This combinatory approach of glycomics and -omics will prove to be crucial in the generation of “designer” biofuels [18].
Additionally, the combination of glycomics and multi-omics provides direct and indirect insights into plant cell wall structure and saccharification of recalcitrant biomass. The use of glycomics in conjugation with -omics has been used to determine the activity and saccharification products of CAZymes in a variety of fields (e.g., human health [219], soil health/carbon sinks [220], novel enzyme discovery [221], and recalcitrant biomass saccharification [222]). However, this strategy is challenged by the complexityof host dynamic microbial ecosystems, CAZymes, and complex carbohydrate structures. Although many researchers have expanded their focus to study CAZymes from anaerobic digesters, leading to an expansion of -omic datasets [157, 177], and likewise, perform glycomic research on biomass saccharification in anaerobic digesters or animal digestive organs [51, 52], there are few studies which combine these tools to fully understand the complexity of anaerobic digesters. Using metatranscriptomics, researchers determined CAZyme expression profiles in Aspergillus niger grown on wheat straw with different pre-treatment methods [223]. The pre-treated wheat straw and resulting growth cultures were analyzed using HPAEC-PAD to determine which CAZymes induced the differential expression patterns between pre-treatment methods. Furthermore, the combination of MAPP, linkage analysis, and metagenomics has recently been used to determine the CAZymes responsible for the digestion of non-soluble polysaccharides in chickens—an approach highly portable to anaerobic digesters [224]. As the field of biofuels progresses, a multi-disciplinary approach will be needed to fine-tune and standardize methods to optimize production, as diversity in microorganisms in combination with feedstocks and feedstock pre-treatments can drastically alter saccharification and fermentation efficiencies.
Conclusion
Improving biofuel production from crop residues is a promising avenue for increasing the value of agricultural waste streams. Although there has been substantial progress made toward understanding the cell wall structure of crop residues, structural variation that exists between plant species and tissues, and chemical modifications resulting from pre-treatments impacts their efficient use in biofuel production. State-of-the-art glycomic methods can be used to provide a high-resolution picture of plant cell wall structure in crop residues, and previous studies have emphasized the importance of using this structural knowledge to detect inefficiencies in biomass fermentation [52, 53] (Fig. 5). Intensified research of crop residue cell wall structure and composition will be informative for designing tailored approaches for individual plant sources. As well, with the advancement of -omics technologies, availability of sequence datasets, and bioinformatic tools developed to interpret metadata, it has become more feasible to discover and deploy novel CAZymes biocatalysts, saccharolytic microbial species, and microbial communities tuned for specific crop residues (Fig. 5). Together, elucidation of biomass cell wall structure and innovations in CAZyme technologies will help streamline future efforts to improve the efficiency of biofuel production, helping unlock the energy potential of agricultural crop waste streams and next-generation biofuel feedstocks.
Availability of data and materials
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Change history
08 February 2021
A Correction to this paper has been published: https://doi.org/10.1186/s13068-021-01888-z
Abbreviations
- CAZyme:
-
Carbohydrate-active enzyme
- PM:
-
Particulate matter
- Kdo:
-
Ketodeoxyoctonic acid
- Dha:
-
3-Deoxy-lyxo-heptulosaric acid
- AX:
-
Arabinoxylan
- GX:
-
Glucuronoxylan
- GlM:
-
Glucomannan
- GaM:
-
Galactomannan
- GGM:
-
Galactoglucomannan
- HG:
-
Homogalacturonan
- RG-I:
-
Rhamnogalacturonan I
- RG-II:
-
Rhamnogalacturonan II
- GAX:
-
Glucuronoarabinoxylan
- HPAEC-PAD:
-
High-performance anion-exchange chromatography with pulsed amperometric detection
- TFA:
-
Trifluoroacetic acid
- RP-HPLC–UV:
-
Reverse-phase high-performance liquid chromatography coupled to ultraviolet detection
- GC–MS/FID:
-
Gas chromatography–mass spectrometry/flame ionization detection
- PMAA:
-
Partially methylated alditol acetates
- LC–ESI–MS/MS:
-
Liquid chromatography electrospray ionization tandem mass spectrometry
- NMR:
-
Nuclear magnetic resonance
- MAPP:
-
Microarray polymer profiling
- mAb:
-
Monoclonal antibodies
- CBM:
-
Carbohydrate-binding module
- GH:
-
Glycoside hydrolase
- PL:
-
Polysaccharide lyase
- CE:
-
Carbohydrate esterase
- AA:
-
Auxiliary activities
- EC:
-
Enzyme class
- LPMO:
-
Lytic polysaccharide monooxygenase
- GT:
-
Glycosyltransferase
- PUL:
-
Polysaccharide Utilization Loci
- HMM:
-
Hidden Markov model
- SACCHARIS:
-
Sequence Analysis and Clustering of CarboHydrate Active enzymes for Rapid Informed prediction of Specificity
- CUPP:
-
The Conserved Unique Peptide Pattern
References
Callegari A, Bolognesi S, Cecconet D, Capodaglio AG. Production technologies, current role, and future prospects of biofuels feedstocks: a state-of-the-art review. Crit Rev Environ Sci Technol. 2020;50(4):384–436.
Campanaro S, Treu L, Kougias PG, De Francisci D, Valle G, Angelidaki I. Metagenomic analysis and functional characterization of the biogas microbiome using high throughput shotgun sequencing and a novel binning strategy. Biotechnol Biofuels. 2016;9(1):26.
Tomei J, Helliwell R. Food versus fuel? Going beyond biofuels. Land Use Policy. 2016;56:320–6.
Lal R. World crop residues production and implications of its use as a biofuel. Environ Int. 2005;31(4):575–84.
Bedoić R, Ćosić B, Duić N. Technical potential and geographic distribution of agricultural residues, co-products and by-products in the European Union. Sci Total Environ. 2019;686:568–79.
Ji L. An assessment of agricultural residue resources for liquid biofuel production in China. Renew Sust Energ Rev. 2015;44:561–75.
Bentsen NS, Felby C, Thorsen BJ. Agricultural residue production and potentials for energy and materials services. Prog Energy Combust Sci. 2014;40:59–73.
Li X, Mupondwa E, Panigrahi S, Tabil L, Sokhansanj S, Stumborg M. A review of agricultural crop residue supply in Canada for cellulosic ethanol production. Renew Sust Energ Rev. 2012;16(5):2954–65.
Haq Z, Easterly JL. Agricultural residue availability in the United States. Appl Biochem Biotechnol. 2006;129(1):3–21.
García-Condado S, López-Lozano R, Panarello L, Cerrani I, Nisini L, Zucchini A, et al. Assessing lignocellulosic biomass production from crop residues in the European Union: Modelling, analysis of the current scenario and drivers of interannual variability. GCB Bioenergy. 2019;11(6):809–31.
Searle SY, Malins CJ. Waste and residue availability for advanced biofuel production in EU Member States. Biomass Bioenerg. 2016;89:2–10.
Ronzon T, Piotrowski S. Are primary agricultural residues promising feedstock for the European bioeconomy? Ind Biotechnol. 2017;13(3):113–27.
Smil V. Crop residues: Agriculture’s largest harvest: Crop residues incorporate more than half of the world’s agricultural phytomass. Bioscience. 1999;49(4):299–308.
Bhuvaneshwari S, Hettiarachchi H, Meegoda JN. Crop residue burning in India: policy challenges and potential solutions. Int J Environ Res Public Health. 2019;16(5):832.
Shi T, Liu Y, Zhang L, Hao L, Gao Z. Burning in agricultural landscapes: an emerging natural and human issue in China. Landsc Ecol. 2014;29(10):1785–98.
Zabed H, Sahu JN, Suely A, Boyce AN, Faruq G. Bioethanol production from renewable sources: current perspectives and technological progress. Renew Sust Energ Rev. 2017;71:475–501.
Terrett OM, Dupree P. Covalent interactions between lignin and hemicelluloses in plant secondary cell walls. Curr Opin Biotech. 2019;56:97–104.
Smith PJ, Wang HT, York WS, Pena MJ, Urbanowicz BR. Designer biomass for next-generation biorefineries: leveraging recent insights into xylan structure and biosynthesis. Biotechnol Biofuels. 2017;10:286.
Olofsson K, Bertilsson M, Lidén G. A short review on SSF—an interesting process option for ethanol production from lignocellulosic feedstocks. Biotechnol Biofuels. 2008;1(1):7.
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(Database issue):D490–5.
Anderson CT, Kieber JJ. Dynamic construction, perception, and remodeling of plant cell walls. Annu Rev Plant Biol. 2020;71(1):39–69.
Pettolino FA, Walsh C, Fincher GB, Bacic A. Determining the polysaccharide composition of plant cell walls. Nat Protoc. 2012;7(9):1590–607.
Chen X, Kim J. Callose synthesis in higher plants. Plant Signal Behav. 2009;4(6):489–92.
Ndeh D, Rogowski A, Cartmell A, Luis AS, Baslé A, Gray J, et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature. 2017;544(7648):65–70.
Gu J, Catchmark JM. The impact of cellulose structure on binding interactions with hemicellulose and pectin. Cellulose. 2013;20(4):1613–27.
O’Neill MA, Warrenfeltz D, Kates K, Pellerin P, Doco T, Darvill AG, et al. Rhamnogalacturonan-II, a pectic polysaccharide in the walls of growing plant cell, forms a dimer that is covalently cross-linked by a borate ester. In vitro conditions for the formation and hydrolysis of the dimer. J Biol Chem. 1996;271(37):22923–30.
Oosterveld A, Grabber JH, Beldman G, Ralph J, Voragen AGJ. Formation of ferulic acid dehydrodimers through oxidative cross-linking of sugar beet pectin. Carbohydr Res. 1997;300(2):179–81.
Agger J, Viksø-Nielsen A, Meyer AS. Enzymatic xylose release from pretreated corn bran arabinoxylan: differential effects of deacetylation and deferuloylation on insoluble and soluble substrate fractions. J Agr Food Chem. 2010;58(10):6141–8.
Scheller HV, Ulvskov P. Hemicelluloses. Annu Rev Plant Biol. 2010;61(1):263–89.
Yokoyama R. A genomic perspective on the evolutionary diversity of the plant cell wall. Plants. 2020;9(9):1195.
Vogel J. Unique aspects of the grass cell wall. Curr Opin Plant Biol. 2008;11(3):301–7.
Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3(1):1–30.
Carpita NC. Structural and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol. 1996;47(1):445–76.
Pattathil S, Hahn MG, Dale BE, Chundawat SPS. Insights into plant cell wall structure, architecture, and integrity using glycome profiling of native and AFEXTM-pre-treated biomass. J Exp Bot. 2015;66(14):4279–94.
Mitchell RAC, Dupree P, Shewry PR. A novel bioinformatics approach identifies candidate genes for the synthesis and feruloylation of arabinoxylan. Plant Physiol. 2007;144(1):43–53.
Ishii T. Structure and functions of feruloylated polysaccharides. Plant Sci. 1997;127(2):111–27.
Ebringerová A, Hromádková Z, Heinze T. Hemicellulose. In: Heinze T, editor. Polysaccharides. Berlin, Heidelberg: Springer; 2005.
Hatfield RD, Wilson JR, Mertens DR. Composition of cell walls isolated from cell types of grain sorghum stems. J Sci Food Agric. 1999;79(6):891–9.
O’Neill MA, York WS. The composition and structure of plant primary cell walls. In: Robert JA, editor. Annual plant reviews. Boca Raton, FL: CRC Press; 2003. p. 1–54.
Grantham NJ, Wurman-Rodrich J, Terrett OM, Lyczakowski JJ, Stott K, Iuga D, et al. An even pattern of xylan substitution is critical for interaction with cellulose in plant cell walls. Nat Plants. 2017;3(11):859–65.
Pustjens AM, Schols HA, Kabel MA, Gruppen H. Characterisation of cell wall polysaccharides from rapeseed (Brassica napus) meal. Carbohydr Polym. 2013;98(2):1650–6.
Wang J, Bai J, Fan M, Li T, Li Y, Qian H, et al. Cereal-derived arabinoxylans: structural features and structure–activity correlations. Trends Food Sci Tech. 2020;96:157–65.
Knudsen KEB. Fiber and nonstarch polysaccharide content and variation in common crops used in broiler diets1. Poult Sci. 2014;93(9):2380–93.
Malunga LN, Beta T. Isolation and identification of feruloylated arabinoxylan mono- and oligosaccharides from undigested and digested maize and wheat. Heliyon. 2016;2(5):e00106.
Huisman MMH, Schols HA, Voragen AGJ. Glucuronoarabinoxylans from maize kernel cell walls are more complex than those from sorghum kernel cell walls. Carbohydr Polym. 2000;43(3):269–79.
Muszyński A, O’Neill MA, Ramasamy E, Pattathil S, Avci U, Peña MJ, et al. Xyloglucan, galactomannan, glucuronoxylan, and rhamnogalacturonan I do not have identical structures in soybean root and root hair cell walls. Planta. 2015;242(5):1123–38.
Morais de Carvalho D, Martínez-Abad A, Evtuguin DV, Colodette JL, Lindström ME, Vilaplana F, et al. Isolation and characterization of acetylated glucuronoarabinoxylan from sugarcane bagasse and straw. Carbohydr Polym. 2017;156:223–34.
Liu L, Paulitz J, Pauly M. The presence of fucogalactoxyloglucan and its synthesis in rice indicates conserved functional importance in plants. Plant Physiol. 2015;168(2):549–60.
Foston M, Samuel R, He J, Ragauskas AJ. A review of whole cell wall NMR by the direct-dissolution of biomass. Green Chem. 2016;18(3):608–21.
Bento-Silva A, Vaz Patto MC, do Rosário BM. Relevance, structure and analysis of ferulic acid in maize cell walls. Food Chem. 2018;246:360–78.
Ke J, Laskar DD, Singh D, Chen S. In situ lignocellulosic unlocking mechanism for carbohydrate hydrolysis in termites: crucial lignin modification. Biotechnol Biofuels. 2011;4(1):17.
Shakeri Yekta S, Hedenström M, Svensson BH, Sundgren I, Dario M, Enrich-Prast A, et al. Molecular characterization of particulate organic matter in full scale anaerobic digesters: an NMR spectroscopy study. Sci Total Environ. 2019;685:1107–15.
Mulat DG, Dibdiakova J, Horn SJ. Microbial biogas production from hydrolysis lignin: insight into lignin structural changes. Biotechnol Biofuels. 2018;11(1):61.
DuBois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28(3):350–6.
Blumenkrantz N, Asboe-Hansen G. New method for quantitative determination of uronic acids. Anal Biochem. 1973;54(2):484–9.
Filisetti-Cozzi TMCC, Carpita NC. Measurement of uronic acids without interference from neutral sugars. Anal Biochem. 1991;197(1):157–62.
Foster CE, Martin TM, Pauly M. Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) Part I: Lignin. J Vis Exp. 2010;37:e1745.
Garcia R, Rakotozafy L, Telef N, Potus J, Nicolas J. Oxidation of ferulic acid or arabinose-esterified ferulic acid by wheat germ peroxidase. J Agr Food Chem. 2002;50(11):3290–8.
Tee-ngam P, Nunant N, Rattanarat P, Siangproh W, Chailapakul O. Simple and rapid determination of ferulic acid levels in food and cosmetic samples using paper-based platforms. Sensors. 2013;13(10):13039–53.
Hestrin S. The reaction of acetylcholine and other carboxylic acid derivatives with hydroxylamine, and its analytical application. J Biol Chem. 1949;180(1):249–61.
Cataldi TRI, Campa C, De Benedetto GE. Carbohydrate analysis by high-performance anion-exchange chromatography with pulsed amperometric detection: the potential is still growing. Fresenius J Anal Chem. 2000;368(8):739–58.
Li J, Wang D, Xing X, Cheng TJR, Liang PH, Bulone V, et al. Structural analysis and biological activity of cell wall polysaccharides extracted from Panax ginseng marc. Int J Biol Macromol. 2019;135:29–37.
De Ruiter GA, Schols HA, Voragen AGJ, Rombouts FM. Carbohydrate analysis of water-soluble uronic acid-containing polysaccharides with high-performance anion-exchange chromatography using methanolysis combined with TFA hydrolysis is superior to four other methods. Anal Biochem. 1992;207(1):176–85.
Willför S, Pranovich A, Tamminen T, Puls J, Laine C, Suurnäkki A, et al. Carbohydrate analysis of plant materials with uronic acid-containing polysaccharides–a comparison between different hydrolysis and subsequent chromatographic analytical techniques. Ind Crops Prod. 2009;29(2):571–80.
Hase S. Chapter 15 pre- and post-column detection-oriented derivatization techniques in HPLC of carbohydrates. In: El Rassi Z, editor. Journal of Chromatography Library. New York: Elsevier; 1995. p. 555–75.
Dai J, Wu Y, Chen S-W, Zhu S, Yin H-P, Wang M, et al. Sugar compositional determination of polysaccharides from Dunaliella salina by modified RP-HPLC method of precolumn derivatization with 1-phenyl-3-methyl-5-pyrazolone. Carbohydr Polym. 2010;82(3):629–35.
Little A, Lahnstein J, Jeffery DW, Khor SF, Schwerdt JG, Shirley NJ, et al. A novel (1,4)-β-linked glucoxylan is synthesized by members of the cellulose synthase-like F gene family in land plants. ACS Cent Sci. 2019;5(1):73–84.
Xing X, Hsieh YSY, Yap K, Ang ME, Lahnstein J, Tucker MR, et al. Isolation and structural elucidation by 2D NMR of planteose, a major oligosaccharide in the mucilage of chia (Salvia hispanica L.) seeds. Carbohydr Polym. 2017;175:231–40.
Ruiz-Matute AI, Hernández-Hernández O, Rodríguez-Sánchez S, Sanz ML, Martínez-Castro I. Derivatization of carbohydrates for GC and GC–MS analyses. J Chromatogr B. 2011;879(17):1226–40.
Sims IM, Carnachan SM, Bell TJ, Hinkley SFR. Methylation analysis of polysaccharides: technical advice. Carbohydr Polym. 2018;188:1–7.
Black I, Heiss C, Azadi P. Comprehensive monosaccharide composition analysis of insoluble polysaccharides by permethylation to produce methyl alditol derivatives for gas chromatography/mass spectrometry. Anal Chem. 2019;91(21):13787–93.
Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 1984;131(2):209–17.
Carpita NC, Shea EM. Linkage structure of carbohydrates by gas chromatography-mass spectrometry (GC-MS) of partially methylated alditol acetates. In: Biermann CJ, McGinnis GD, editors. Analysis of carbohydrates by GLC and MS. Boca Raton, Florida: CRC Press, Inc.; 1989. p. 157–216.
Kim JB, Carpita NC. Changes in esterification of the uronic acid groups of cell wall polysaccharides during elongation of maize coleoptiles. Plant Physiol. 1992;98(2):646–53.
Bacic A, Moody SF, Clarke AE. Structural analysis of secreted root slime from maize (Zea mays L.). Plant Physiol. 1986;80(3):771–7.
Bac VH, Paulsen BS, Truong LV, Koschella A, Trinh TC, Wold CW, et al. Neutral polysaccharide from the leaves of Pseuderanthemum carruthersii: presence of 3-O-methyl galactose and anti-inflammatory activity in LPS-stimulated RAW 2647 cells. Polymers. 2019;11(7):1219.
Carpita NC, Whittern D. A highly substituted glucuronoarabinoxylan from developing maize coleoptiles. Carbohydr Res. 1986;146(1):129–40.
Chiovitti A, Bacic A, Craik DJ, Kraft GT, Liao M-L. A nearly idealized 6′-O-methylated η-carrageenan from the Australian red alga Claviclonium ovatum (Acrotylaceae, Gigartinales). Carbohydr Res. 2004;339(8):1459–66.
John HP. Neutral polysaccharides. In: Chaplin MF, Kennedy JF, editors. Carbohydrate analysis A practical approach (second edition). Oxford: Oxford University Press; 1994.
Jones DR, Xing X, Tingley JP, Klassen L, King ML, Alexander TW, et al. Analysis of active site architecture and reaction product linkage chemistry reveals a conserved cleavage substrate for an endo-alpha-mannanase within diverse yeast mannans. J Mol Biol. 2020;432(4):1083–97.
Huang YL, Jhou BY, Chen SF, Khoo KH. Identifying specific and differentially linked glycosyl residues in mammalian glycans by targeted LC-MS analysis. Anal Sci. 2018;34(9):1049–54.
Galermo AG, Nandita E, Barboza M, Amicucci MJ, Vo T-TT, Lebrilla CB. Liquid chromatography-tandem mass spectrometry approach for determining glycosidic linkages. Anal Chem. 2018;90(21):13073–80.
Galermo AG, Nandita E, Castillo JJ, Amicucci MJ, Lebrilla CB. Development of an extensive linkage library for characterization of carbohydrates. Anal Chem. 2019;91(20):13022–31.
Amicucci MJ, Galermo AG, Guerrero A, Treves G, Nandita E, Kailemia MJ, et al. Strategy for structural elucidation of polysaccharides: elucidation of a maize mucilage that harbors diazotrophic bacteria. Anal Chem. 2019;91(11):7254–65.
Alagesan K, Silva DV, Seeberger PH, Kolarich D. A novel, ultrasensitive approach for quantitative carbohydrate composition and linkage analysis using LC-ESI ion trap tandem mass spectrometry. bioRxiv. 2019. Doi:https://doi.org/10.1101/853036v1.
Yang H, Shi L, Zhuang X, Su R, Wan D, Song F, et al. Identification of structurally closely related monosaccharide and disaccharide isomers by PMP labeling in conjunction with IM-MS/MS. Sci Rep. 2016;6(1):28079.
Wu X, Jiang W, Lu J, Yu Y, Wu B. Analysis of the monosaccharide composition of water-soluble polysaccharides from Sargassum fusiforme by high performance liquid chromatography/electrospray ionisation mass spectrometry. Food Chem. 2014;145:976–83.
Guo N, Bai Z, Jia W, Sun J, Wang W, Chen S, et al. Quantitative analysis of polysaccharide composition in polyporus umbellatus by HPLC-ESI-TOF-MS. Molecules. 2019;24(14):2526.
Cheng HN, Neiss TG. Solution NMR spectroscopy of food polysaccharides. Polym Rev. 2012;52(2):81–114.
Liu J, Zhao Y, Wu Q, John A, Jiang Y, Yang J, et al. Structure characterisation of polysaccharides in vegetable “okra” and evaluation of hypoglycemic activity. Food Chem. 2018;242:211–6.
Ndukwe IE, Black I, Heiss C, Azadi P. Evaluating the utility of permethylated polysaccharides. Solution NMR data for characterization of insoluble plant cell wall polysaccharides. Anal Chem. 2020;92:13221.
Kim H, Ralph J. Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6/pyridine-d5. Org Biomol Chem. 2010;8(3):576–91.
Yelle DJ, Ralph J, Frihart CR. Characterization of nonderivatized plant cell walls using high-resolution solution-state NMR spectroscopy. Magn Reson Chem. 2008;46(6):508–17.
Kim H, Ralph J, Akiyama T. Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6. BioEnergy Res. 2008;1(1):56–66.
Mansfield SD, Kim H, Lu F, Ralph J. Whole plant cell wall characterization using solution-state 2D NMR. Nat Protoc. 2012;7(9):1579–89.
Kirui A, Dickwella Widanage MC, Mentink-Vigier F, Wang P, Kang X, Wang T. Preparation of fungal and plant materials for structural elucidation using dynamic nuclear polarization solid-state NMR. J Vis Exp. 2019;144:e59152.
Zhao W, Fernando LD, Kirui A, Deligey F, Wang T. Solid-state NMR of plant and fungal cell walls: A critical review. Solid State Nucl Magn Reson. 2020;107:101660.
Kang X, Kirui A, Dickwella Widanage MC, Mentink-Vigier F, Cosgrove DJ, Wang T. Lignin-polysaccharide interactions in plant secondary cell walls revealed by solid-state NMR. Nat Commun. 2019;10(1):347.
Pattathil S, Avci U, Miller JS, Hahn MG. Immunological approaches to plant cell wall and biomass characterization: glycome profiling. In: Himmel M, editor. Biomass Conversion Methods in Molecular Biology (Methods and Protocols). Totowa, NJ: Humana Press; 2012.
Moller I, Sørensen I, Bernal AJ, Blaukopf C, Lee K, Øbro J, et al. High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J. 2007;50(6):1118–28.
DeMartini JD, Pattathil S, Avci U, Szekalski K, Mazumder K, Hahn MG, et al. Application of monoclonal antibodies to investigate plant cell wall deconstruction for biofuels production. Energy Environ Sci. 2011;4(10):4332–9.
Kataeva I, Foston MB, Yang S-J, Pattathil S, Biswal AK, Poole Ii FL, et al. Carbohydrate and lignin are simultaneously solubilized from unpretreated switchgrass by microbial action at high temperature. Energy Environ Sci. 2013;6(7):2186–95.
Gao Y, Fangel JU, Willats WGT, Moore JP. Tracking polysaccharides during white winemaking using glycan microarrays reveals glycoprotein-rich sediments. Food Res Int. 2019;123:662–73.
Fangel JU, Eiken J, Sierksma A, Schols HA, Willats WGT, Harholt J. Tracking polysaccharides through the brewing process. Carbohydr Polym. 2018;196:465–73.
Ahl LI, Grace OM, Pedersen HL, Willats WGT, Jørgensen B, Rønsted N. Analyses of Aloe polysaccharides using carbohydrate microarray profiling. J AOAC Int. 2019;101(6):1720–8.
Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3(9):853–9.
Davies GJ, Henrissat B. Structural enzymology of carbohydrate-active enzymes: implications for the post-genomic era. Biochem Soc Trans. 2002;30(2):291–7.
Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM, Henrissat B. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J. 2010;432(3):437–44.
Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels. 2013;6(1):41.
Davies GJ, Gloster TM, Henrissat B. Recent structural insights into the expanding world of carbohydrate-active enzymes. Curr Opin Struct Biol. 2005;15(6):637–45.
Lapébie P, Lombard V, Drula E, Terrapon N, Henrissat B. Bacteroidetes use thousands of enzyme combinations to break down glycans. Nat Commun. 2019;10(1):2043.
Asatsuma S, Sawada C, Itoh K, Okito M, Kitajima A, Mitsui T. Involvement of alpha-amylase I-1 in starch degradation in rice chloroplasts. Plant Cell Physiol. 2005;46(6):858–69.
Payne CM, Knott BC, Mayes HB, Hansson H, Himmel ME, Sandgren M, et al. Fungal cellulases. Chem Rev. 2015;115(3):1308–448.
Luis AS, Briggs J, Zhang X, Farnell B, Ndeh D, Labourel A, et al. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol. 2018;3(2):210–9.
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–5.
Garron ML, Cygler M. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology. 2010;20(12):1547–73.
Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sørlie M, et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330(6001):219–22.
Vermaas JV, Crowley MF, Beckham GT, Payne CM. Effects of lytic polysaccharide monooxygenase oxidation on cellulose structure and binding of oxidized cellulose oligomers to cellulases. J Phys Chem B. 2015;119(20):6129–43.
Hemsworth GR, Johnston EM, Davies GJ, Walton PH. Lytic polysaccharide monooxygenases in biomass conversion. Trends Biotechnol. 2015;33(12):747–61.
Arantes V, Saddler JN. Access to cellulose limits the efficiency of enzymatic hydrolysis: the role of amorphogenesis. Biotechnol Biofuels. 2010;3:4.
Henrissat B, Driguez H, Viet C, Schülein M. Synergism of cellulases from Trichoderma reesei in the degradation of cellulose. Bio/Technology. 1985;3(8):722–6.
Ma L, Zhang J, Zou G, Wang C, Zhou Z. Improvement of cellulase activity in Trichoderma reesei by heterologous expression of a beta-glucosidase gene from Penicillium decumbens. Enzyme Microb Technol. 2011;49(4):366–71.
Ezeilo UR, Zakaria II, Huyop F, Wahab RA. Enzymatic breakdown of lignocellulosic biomass: the role of glycosyl hydrolases and lytic polysaccharide monooxygenases. Biotechnol Biotechnol Equip. 2017:1–16.
Gilbert HJ, Hazlewood GP. Bacterial cellulases and xylanases. Microbiology. 1993;139(2):187–94.
Saini JK, Saini R, Tewari L. Lignocellulosic agriculture wastes as biomass feedstocks for second-generation bioethanol production: concepts and recent developments. Biotech. 2015;5(4):337–53.
Xiros C, Topakas E, Christakopoulos P. Hydrolysis and fermentation for cellulosic ethanol production. WIRE Energy Environ. 2013;2(6):633–54.
Yeoman CJ, Han Y, Dodd D, Schroeder CM, Mackie RI, Cann IKO. Thermostable enzymes as biocatalysts in the biofuel industry. Adv Appl Microbiol. 2010;70:1–55.
Srivastava N, Mishra PK, Upadhyay SN. Endoglucanase: revealing participation in open cellulosic chains. In: Srivastava N, Mishra PK, Upadhyay SN, editors. Industrial Enzymes for Biofuels Production. Amsterdam: Elsevier; 2020. p. 37–62.
Vlasenko E, Schülein M, Cherry J, Xu F. Substrate specificity of family 5, 6, 7, 9, 12, and 45 endoglucanases. Bioresour Technol. 2010;101(7):2405–11.
Aspeborg H, Coutinho PM, Wang Y, Brumer H, Henrissat B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol Biol. 2012;12(1):186.
Frommhagen M, Westphal AH, van Berkel WJH, Kabel MA. Distinct substrate specificities and electron-donating systems of fungal lytic polysaccharide monooxygenases. Front Microbiol. 2018;9:1080.
Srivastava N, Mishra PK, Upadhyay SN. Xylanases: For digestion of hemicellulose. In: Srivastava N, Mishra PK, Upadhyay SN, editors. Industrial enzymes for biofuels production. Amsterdam: Elsevier; 2020. p. 101–32.
Beaugrand J, Chambat G, Wong VWK, Goubet F, Rémond C, Paës G, et al. Impact and efficiency of GH10 and GH11 thermostable endoxylanases on wheat bran and alkali-extractable arabinoxylans. Carbohydr Res. 2004;339(15):2529–40.
Mewis K, Lenfant N, Lombard V, Henrissat B. Dividing the large glycoside hydrolase family 43 into subfamilies: a motivation for detailed enzyme characterization. Appl Environ Microbiol. 2016;82(6):1686–92.
Hagen LH, Brooke CG, Shaw CA, Norbeck AD, Piao H, Arntzen MØ, et al. Proteome specialization of anaerobic fungi during ruminal degradation of recalcitrant plant fiber. ISME J. 2020. https://doi.org/10.1038/s41396-020-00769-x.
Faulds CB, Kroon PA, Saulnier L, Thibault J-F, Williamson G. Release of ferulic acid from maize bran and derived oligosaccharides by Aspergillus niger esterases. Carbohydr Polym. 1995;27(3):187–90.
Saulnier L, Marot C, Elgorriaga M, Bonnin E, Thibault JF. Thermal and enzymatic treatments for the release of free ferulic acid from maize bran. Carbohydr Polym. 2001;45(3):269–75.
Grabber JH, Ralph J, Hatfield RD. Ferulate cross-links limit the enzymatic degradation of synthetically lignified primary walls of maize. J Agr Food Chem. 1998;46(7):2609–14.
Agger JW, Isaksen T, Várnai A, Vidal-Melgosa S, Willats WGT, Ludwig R, et al. Discovery of LPMO activity on hemicelluloses shows the importance of oxidative processes in plant cell wall degradation. PNAS. 2014;111(17):6287–92.
Campbell JA, Davies GJ, Bulone VV, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1998;329(Pt 3):719.
Keegstra K, Raikhel N. Plant glycosyltransferases. Curr Opin Plant Biol. 2001;4(3):219–24.
Sticklen MB. Plant genetic engineering for biofuel production: towards affordable cellulosic ethanol. Nat Rev Genet. 2008;9(6):433–43.
Biswal AK, Atmodjo MA, Li M, Baxter HL, Yoo CG, Pu Y, et al. Sugar release and growth of biofuel crops are improved by downregulation of pectin biosynthesis. Nat Biotechnol. 2018;36(3):249–57.
Li M, Yoo CG, Pu Y, Biswal AK, Tolbert AK, Mohnen D, et al. Downregulation of pectin biosynthesis gene GAUT4 leads to reduced ferulate and lignin-carbohydrate cross-linking in switchgrass. Commun Biol. 2019;2:22.
Srivastava N, Mishra PK, Upadhyay SN. Laccase: use in removal of lignin in cellulosic biomass. In: Srivastava N, Mishra PK, Upadhyay SN, editors. Industrial enzymes for biofuels production. Amsterdam: Elsevier; 2020. p. 133–57.
Arnling Baath J, Mazurkewich S, Knudsen RM, Poulsen JN, Olsson L, Lo Leggio L, et al. Biochemical and structural features of diverse bacterial glucuronoyl esterases facilitating recalcitrant biomass conversion. Biotechnol Biofuels. 2018;11:213.
Adsul M, Sandhu SK, Singhania RR, Gupta R, Puri SK, Mathur A. Designing a cellulolytic enzyme cocktail for the efficient and economical conversion of lignocellulosic biomass to biofuels. Enzyme Microb Technol. 2020;133:109442.
Kudanga T, Le Roes-Hill M. Laccase applications in biofuels production: current status and future prospects. Appl Microbiol Biotechnol. 2014;98(15):6525–42.
Murray PG, Grassick A, Laffey CD, Cuffe MM, Higgins T, Savage AV, et al. Isolation and characterization of a thermostable endo-β-glucanase active on 1,3–1,4-β-d-glucans from the aerobic fungus Talaromyces emersonii CBS 814.70. Enzyme Microb Technol. 2001;29(1):90–8.
Szijártó N, Siika-aho M, Tenkanen M, Alapuranen M, Vehmaanperä J, Réczey K, et al. Hydrolysis of amorphous and crystalline cellulose by heterologously produced cellulases of Melanocarpus albomyces. J Biotechnol. 2008;136(3):140–7.
Gupta VK, Kubicek CP, Berrin J-G, Wilson DW, Couturier M, Berlin A, et al. Fungal enzymes for bio-products from sustainable and waste biomass. Trends Biochem Sci. 2016;41(7):633–45.
Binod P, Gnansounou E, Sindhu R, Pandey A. Enzymes for second generation biofuels: recent developments and future perspectives. Bioresour Technol rep. 2019;5:317–25.
Akinosho H, Yee K, Close D, Ragauskas A. The emergence of Clostridium thermocellum as a high utility candidate for consolidated bioprocessing applications. Front Chem. 2014;2:66.
Tanghe M, Danneels B, Camattari A, Glieder A, Vandenberghe I, Devreese B, et al. Recombinant expression of Trichoderma reesei Cel61A in Pichia pastoris: optimizing yield and N-terminal processing. Mol Biotechnol. 2015;57(11):1010–7.
Verastegui Y, Cheng J, Engel K, Kolczynski D, Mortimer S, Lavigne J, et al. Multisubstrate isotope labeling and metagenomic analysis of active soil bacterial communities. mBio. 2014;5(4):e01157-e1214.
Wang C, Dong D, Wang H, Müller K, Qin Y, Wang H, et al. Metagenomic analysis of microbial consortia enriched from compost: new insights into the role of Actinobacteria in lignocellulose decomposition. Biotechnol Biofuels. 2016;9(1):22.
Wilkens C, Busk PK, Pilgaard B, Zhang W-J, Nielsen KL, Nielsen PH, et al. Diversity of microbial carbohydrate-active enzymes in Danish anaerobic digesters fed with wastewater treatment sludge. Biotechnol Biofuels. 2017;10(1):158.
Hess M, Sczyrba A, Egan R, Kim T-W, Chokhawala H, Schroth G, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331(6016):463–7.
Romero Victorica M, Soria MA, Batista-García RA, Ceja-Navarro JA, Vikram S, Ortiz M, et al. Neotropical termite microbiomes as sources of novel plant cell wall degrading enzymes. Sci Rep. 2020;10(1):3864.
Vinzelj J, Joshi A, Insam H, Podmirseg SM. Employing anaerobic fungi in biogas production: challenges & opportunities. Bioresour Technol. 2020;300:122687.
Li H, Yelle DJ, Li C, Yang M, Ke J, Zhang R, et al. Lignocellulose pretreatment in a fungus-cultivating termite. PNAS. 2017;114(18):4709.
Zhu N, Yang J, Ji L, Liu J, Yang Y, Yuan H. Metagenomic and metaproteomic analyses of a corn stover-adapted microbial consortium EMSD5 reveal its taxonomic and enzymatic basis for degrading lignocellulose. Biotechnol Biofuels. 2016;9(1):243.
Maus I, Koeck DE, Cibis KG, Hahnke S, Kim YS, Langer T, et al. Unraveling the microbiome of a thermophilic biogas plant by metagenome and metatranscriptome analysis complemented by characterization of bacterial and archaeal isolates. Biotechnol Biofuels. 2016;9(1):171.
Ye J, Li D, Sun Y, Wang G, Yuan Z, Zhen F, et al. Improved biogas production from rice straw by co-digestion with kitchen waste and pig manure. Waste Manage. 2013;33(12):2653–8.
Ozbayram EG, Kleinsteuber S, Nikolausz M, Ince B, Ince O. Effect of bioaugmentation by cellulolytic bacteria enriched from sheep rumen on methane production from wheat straw. Anaerobe. 2017;46:122–30.
Güllert S, Fischer MA, Turaev D, Noebauer B, Ilmberger N, Wemheuer B, et al. Deep metagenome and metatranscriptome analyses of microbial communities affiliated with an industrial biogas fermenter, a cow rumen, and elephant feces reveal major differences in carbohydrate hydrolysis strategies. Biotechnol Biofuels. 2016;9(1):121.
Stewart EJ. Growing unculturable bacteria. J Bacteriol Res. 2012;194(16):4151.
Xia Y, Wang Y, Wang Y, Chin FYL, Zhang T. Cellular adhesiveness and cellulolytic capacity in Anaerolineae revealed by omics-based genome interpretation. Biotechnol Biofuels. 2016;9(1):111.
Stewart RD, Auffret MD, Warr A, Walker AW, Roehe R, Watson M. Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat Biotechnol. 2019;37(8):953–61.
Xia Y, Ju F, Fang HHP, Zhang T. Mining of novel thermo-stable cellulolytic genes from a thermophilic cellulose-degrading consortium by metagenomics. PLoS ONE. 2013;8(1):e53779.
Bremges A, Maus I, Belmann P, Eikmeyer F, Winkler A, Albersmeier A, et al. Deeply sequenced metagenome and metatranscriptome of a biogas-producing microbial community from an agricultural production-scale biogas plant. GigaScience. 2015;4(1):33.
Abendroth C, Simeonov C, Peretó J, Antúnez O, Gavidia R, Luschnig O, et al. From grass to gas: microbiome dynamics of grass biomass acidification under mesophilic and thermophilic temperatures. Biotechnol Biofuels. 2017;10(1):171.
Heyer R, Benndorf D, Kohrs F, De Vrieze J, Boon N, Hoffmann M, et al. Proteotyping of biogas plant microbiomes separates biogas plants according to process temperature and reactor type. Biotechnol Biofuels. 2016;9(1):155.
Vanwonterghem I, Jensen PD, Ho DP, Batstone DJ, Tyson GW. Linking microbial community structure, interactions and function in anaerobic digesters using new molecular techniques. Curr Opin Biotech. 2014;27:55–64.
Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol. 2017;35(8):725–31.
Frioux C, Singh D, Korcsmaros T, Hildebrand F. From bag-of-genes to bag-of-genomes: metabolic modelling of communities in the era of metagenome-assembled genomes. Comput Struct Biotechnol J. 2020;18:1722–34.
Campanaro S, Treu L, Rodriguez-R LM, Kovalovszki A, Ziels RM, Maus I, et al. New insights from the biogas microbiome by comprehensive genome-resolved metagenomics of nearly 1600 species originating from multiple anaerobic digesters. Biotechnol Biofuels. 2020;13(1):25.
West P, Probst A, Grigoriev I, Thomas B, Banfield J. Genome-reconstruction for eukaryotes from complex natural microbial communities. Genome Res. 2018;28:gr.228429.117.
Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35(9):833–44.
Ju F, Zhang T. Experimental design and bioinformatics analysis for the application of metagenomics in environmental sciences and biotechnology. Environ Sci Technol. 2015;49(21):12628–40.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–9.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9(1):75.
Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinform. 2011;12(1):491.
Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(suppl_2):ii215.
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44(14):6614–24.
Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28(1):33–6.
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 2014;42(2):D206–14.
El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–32.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Consortium TC. Ten years of CAZypedia: a living encyclopedia of carbohydrate-active enzymes. Glycobiology. 2017;28(1):3–8.
Terrapon N, Lombard V, Drula É, Lapébie P, Al-Masaudi S, Gilbert HJ, et al. PULDB: the expanded database of Polysaccharide Utilization Loci. Nucleic Acids Res. 2018;46(D1):D677–83.
Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46(W1):W95–101.
Park BH, Karpinets TV, Syed MH, Leuze MR, Uberbacher EC. CAZymes Analysis Toolkit (CAT): Web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology. 2010;20(12):1574–84.
Nguyen SN, Flores A, Talamantes D, Dar F, Valdez A, Schwans J, et al. GeneHunt for rapid domain-specific annotation of glycoside hydrolases. Sci Rep. 2019;9(1):10137.
Jones DR, Thomas D, Alger N, Ghavidel A, Inglis GD, Abbott DW. SACCHARIS: an automated pipeline to streamline discovery of carbohydrate active enzyme activities within polyspecific families and de novo sequence datasets. Biotechnol Biofuels. 2018;11(1):27.
Barrett K, Lange L. Peptide-based functional annotation of carbohydrate-active enzymes by conserved unique peptide patterns (CUPP). Biotechnol Biofuels. 2019;12(1):102.
Kultima JR, Coelho LP, Forslund K, Huerta-Cepas J, Li SS, Driessen M, et al. MOCAT2: a metagenomic assembly, annotation and profiling framework. Bioinformatics. 2016;32(16):2520–3.
Stam MR, Danchin EGJ, Rancurel C, Coutinho PM, Henrissat B. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Protein Eng Des Sel. 2006;19(12):555–62.
Viborg AH, Terrapon N, Lombard V, Michel G, Czjzek M, Henrissat B, et al. A subfamily roadmap of the evolutionarily diverse glycoside hydrolase family 16 (GH16). J Biol Chem. 2019;294(44):15973–86.
St John FJ, González JM, Pozharski E. Consolidation of glycosyl hydrolase family 30: A dual domain 4/7 hydrolase family consisting of two structurally distinct groups. FEBS Lett. 2010;584(21):4435–41.
Stewart RD, Auffret MD, Roehe R, Watson M. Open prediction of polysaccharide utilisation loci (PUL) in 5414 public Bacteroidetes genomes using PULpy. bioRxiv. 2018. Doi:https://doi.org/10.1101/421024v1.full.
Shaffer M, Borton MA, McGivern BB, Zayed AA, La Rosa SL, Solden LM, et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. bioRxiv. 2020. Doi: https://doi.org/10.1101/2020.06.29.177501v1.full.
Ausland C, Zheng J, Yi H, Yang B, Li T, Feng X, et al. dbCAN-PUL: a database of experimentally characterized CAZyme gene clusters and their substrates. Nucleic Acids Res. 2020.
Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5(1):113.
Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–5.
Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5(3):e9490.
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3.
Jones DR, Uddin MS, Gruninger RJ, Pham TTM, Thomas D, Boraston AB, et al. Discovery and characterization of family 39 glycoside hydrolases from rumen anaerobic fungi with polyspecific activity on rare arabinosyl substrates. J Biol Chem. 2017;292(30):12606–20.
Bhandari P, Tingley JP, Abbott DW, Hill JE. Characterization of an α-glucosidase enzyme conserved in Gardnerella spp. isolated from the human vaginal microbiome. bioRxiv. 2020. Doi: https://doi.org/10.1101/2020.05.11.086124v2.
Barrett K, Hunt CJ, Lange L, Meyer AS. Conserved unique peptide patterns (CUPP) online platform: peptide-based functional annotation of carbohydrate active enzymes. Nucleic Acids Res. 2020;48(W1):W110–5.
Zeuner B, Thomsen TB, Stringer MA, Krogh KBRM, Meyer AS, Holck J. Comparative characterization of aspergillus pectin lyases by discriminative substrate degradation profiling. Front Bioeng Biotechnol. 2020;8:873.
Pilgaard B, Wilkens C, Herbst F-A, Vuillemin M, Rhein-Knudsen N, Meyer AS, et al. Proteomic enzyme analysis of the marine fungus Paradendryphiella salina reveals alginate lyase as a minimal adaptation strategy for brown algae degradation. Sci Rep. 2019;9(1):12338.
Barrett K, Jensen K, Meyer AS, Frisvad JC, Lange L. Fungal secretome profile categorization of CAZymes by function and family corresponds to fungal phylogeny and taxonomy: Example Aspergillus and Penicillium. Sci Rep. 2020;10(1):5158.
Minic Z, Jouanin L. Plant glycoside hydrolases involved in cell wall polysaccharide degradation. Plant Physiol Biochem. 2006;44(7):435–49.
Fangel J, Ulvskov P, Knox JP, Mikkelsen M, Harholt J, Popper Z, et al. Cell wall evolution and diversity. Front Plant Sci. 2012;3:152.
Jensen JK, Busse-Wicher M, Poulsen CP, Fangel JU, Smith PJ, Yang J-Y, et al. Identification of an algal xylan synthase indicates that there is functional orthology between algal and plant cell wall biosynthesis. New Phytol. 2018;218(3):1049–60.
Lunin VV, Wang H-T, Bharadwaj VS, Alahuhta M, Peña MJ, Yang J-Y, et al. Molecular Mechanism of Polysaccharide Acetylation by the Arabidopsis Xylan O-acetyltransferase XOAT1. Plant Cell. 2020;32(7):2367.
Wang X, Tang Q, Zhao X, Jia C, Yang X, He G, et al. Functional conservation and divergence of Miscanthus lutarioriparius GT43 gene family in xylan biosynthesis. BMC Plant Biol. 2016;16(1):102.
Crouch LI, Liberato MV, Urbanowicz PA, Baslé A, Lamb CA, Stewart CJ, et al. Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown. Nat Commun. 2020;11(1):4017.
McKee LS, Martínez-Abad A, Ruthes AC, Vilaplana F, Brumer H. Focused Metabolism of β-Glucans by the Soil Bacteroidetes Species Chitinophaga pinensis. Appl Environ Microbiol. 2019;85(2):e02231-e2318.
Helbert W, Poulet L, Drouillard S, Mathieu S, Loiodice M, Couturier M, et al. Discovery of novel carbohydrate-active enzymes through the rational exploration of the protein sequences space. PNAS. 2019;116(13):6063.
Armstrong Z, Mewis K, Liu F, Morgan-Lang C, Scofield M, Durno E, et al. Metagenomics reveals functional synergy and novel polysaccharide utilization loci in the Castor canadensis fecal microbiome. ISME J. 2018;12(11):2757–69.
Daly P, van Munster JM, Blythe MJ, Ibbett R, Kokolski M, Gaddipati S, et al. Expression of Aspergillus niger CAZymes is determined by compositional changes in wheat straw generated by hydrothermal or ionic liquid pretreatments. Biotechnol Biofuels. 2017;10:35.
Low K, Xing X, Moote P, Inglis G, Venketachalam S, Hahn M, et al. Combinatorial glycomic analyses to direct CAZyme discovery for the tailored degradation of canola meal non-starch dietary polysaccharides. Microorganisms. 2020;8:1888.
Schultink A, Liu L, Zhu L, Pauly M. Structural diversity and function of xyloglucan sidechain substituents. Plants. 2014;3(4):526–42.
Pauly M, Gille S, Liu L, Mansoori N, Souza A, Schultink A, et al. Hemicellulose biosynthesis. Planta. 2013;238:627.
Neelamegham S, Aoki-Kinoshita K, Bolton E, Frank M, Lisacek F, Lütteke T, et al. Updates to the symbol nomenclature for glycans guidelines. Glycobiology. 2019;29(9):620–4.
Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–9.
The PyMOL Molecular Graphics System VS, LLC.
Acknowledgements
N/A.
Funding
This work was supported by funding from Agriculture and Agri-Food Canada (Project No: J-002262 and J-001589). XX is supported by funding from Alberta Agriculture and Forestry (Project no: 2019H001R).
Author information
Authors and Affiliations
Contributions
JPT, KEL, XX, and DWA contributed to the writing and editing of the manuscript. JPT generated figures with feedback from all the authors. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The errors in the figures have been corrected.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tingley, J.P., Low, K.E., Xing, X. et al. Combined whole cell wall analysis and streamlined in silico carbohydrate-active enzyme discovery to improve biocatalytic conversion of agricultural crop residues. Biotechnol Biofuels 14, 16 (2021). https://doi.org/10.1186/s13068-020-01869-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13068-020-01869-8