Abstract
Background
Blastic plasmacytoid dendritic cell neoplasm (BPDCN) is a rare hematologic malignancy. Based on literature reports of limited cases, over 50 % of BPDCN have chromosomal abnormalities, but no single chromosomal change has been identified as diagnostic of this entity.
Case presentation
In this report, we present a case of BPDCN with complicated chromosomal abnormalities involving chromosomes 12 and 22 and resulting in a simultaneous partial deletion of ETV6 and EWSR1. Notably, these aberrations were identified in bone marrow myeloid precursors in the absence of bone marrow involvement by BPDCN.
Conclusion
Analysis of 46 BPDCN cases with abnormal karyotypes (45 from literature reports plus this case) showed that 12p- is one of the most common structural aberrations in BPDCN. The ETV6 and CDKN1B on 12p deserve further investigations as potential markers of BPDCN.
Similar content being viewed by others
Background
Blastic plasmacytoid dendritic cell neoplasm (BPDCN) is a rare, aggressive myeloid neoplasm derived from plasmacytoid dendritic cells [1]. This disease has been previously described in the past using terms such as “acute agranular CD4+ natural killer (NK) cell leukemia” [2], “blastic NK cell lymphoma” [3] and “agranular CD4 + CD56+ hematodermic neoplasm/tumor” [4, 5]. BPDCN may involve multiple sites, commonly skin, bone marrow (BM), peripheral blood (PB) and lymph nodes (LN). Based on literature reports of limited cases, over 50 % of BPDCN have chromosomal abnormalities, but no single chromosomal change has been shown to be diagnostic of this entity. The common chromosomal aberrations in BPDCN reported previously include abnormalities involving chromosomes 5q (72 %), 12p (64 %), 13q (64 %), 6q (50 %), 15q (43 %), and 9 (usually monosomy 9, 28 %) [4, 6]. Aberrations of 12p are among the most common findings in BPDCN.
In this report, we present a case of BPDCN with complicated chromosomal abnormalities involving chromosomes 12 and 22 and resulting in simultaneous partial deletion of ETV6 and EWSR1. These findings were identified in the BM of a patient with BPDCN in the absence of morphologic, immunohistochemical, or flow cytometry evidence of BPDCN. We also conduct a literature review of conventional cytogenetic findings in BPDCN.
Case presentation
The patient is a previously healthy 44-year-old man who presented with a painless enlarging mass in his left groin. He was observed initially for three months and eventually was referred for an excisional lymph node biopsy. Histologic examination showed a high-grade malignant neoplasm that was diagnosed as BPDCN. He was then referred to our institution. BM evaluation included a trephine biopsy and aspiration. There was no evidence of BPDCN in BM by morphology or immunohistochemistry. Flow cytometry was also negative for BPDCN in BM. However, conventional cytogenetic analysis performed on the BM aspirate sample showed karyotypic aberrations involving chromosomes 12 and 22, which were further characterized by fluorescence in situ hybridization (FISH) analysis (see details below). The patient was treated with a hyper-CVAD-Bortezomib regimen (hyperfractioned cyclophosphamide, vincristine, doxorubicin, dexamethasone alternating with high dose of methotrexate and cytarabine, plus bortezomib) regimen. He also received prophylactic intrathecal chemotherapy with methotrexate for 3 cycles and achieved a complete remission.
Methods and results
Conventional chromosomal analysis
Conventional chromosomal analysis (karyotyping) was performed on G-banded metaphase cells prepared from unstimulated 24-h and 48-h bone marrow cultures as described previously [7]. Twenty metaphases (10 from each culture) were analyzed. The chromosomal abnormalities were reported according to the International System for Human Cytogenetic Nomenclature 2013 (ISCN2013) guidelines [8]. Out of 20 metaphases analyzed, 10 exhibited structural abnormalities involving chromosomes 12 and 22.
Fluorescence in situ Hybridization (FISH) analysis
The following FISH probes were applied in this study: Vysis ETV6 Break Apart (BAP) FISH probe and Vysis EWSR1 BAP FISH probe (Abbott Molecular, Des Plaines, IL) were used for interphase, metaphase and tissue FISH tests. The Vysis MYC BAP FISH probe (Abbott Molecular, Des Plaines, IL) was used for both interphase and tissue FISH tests. The Vysis LSI BCR/ABL ES Dual Color Fusion probe (Abbott Molecular, Des Plaines, IL) was used for interphase FISH tests, whereas the Aquarius Whole Chromosome Painting (WCP) probes for chromosomes 12 and 22 (Cytocell, Tarrytown, NY) were used for metaphase FISH only. All probes were thoroughly validated in accordance with the American College of Medical Genetics and Genomics (ACMGG) guidelines.
Interphase FISH performed on unstimulated cultured cells from BM sample using the ETV6 BAP probe showed that about 25 % of cells had a one-red-one-fusion (1R1F) signal pattern, indicating an ETV6 gene rearrangement with a partial deletion of the 3′ETV6 (green signal). A separate interphase FISH analysis using the EWSR1 BAP probe showed that almost the same percentage of cells had a one-green-one-fusion (1G1F) signal pattern, demonstrating an EWSR1 gene rearrangement with a partial deletion of the 5′EWSR1 (red signal). Mapping back to previously G-banded and karyotyped metaphases showed that the existing 5′ETV6 (red signal) is located at the long arm of the abnormal chromosome 22, whereas the existing 3′EWSR1 (green signal) is located at the short arm of the abnormal chromosome 12 (Fig. 1). Whole chromosome painting (WCP) further confirmed the origins as abnormal chromosomes 12 and 22 (images not shown). Therefore, the results of conventional cytogenetic analysis and FISH analysis suggest that a translocation between 12p and 22q occurred. This was likely followed by a pericentric inversion of the abnormal chromosome 12 resulting in a partial deletion of both 3′ETV6 and 5′EWSR1. These two abnormal chromosomes are described as der(12)t(12;22)(p13;q12)del(22)(q12q12)inv(12)(p13q24.1) and der(22)t(12;22)del(12)(p13p13).
Mapping back to G-banded Metaphases with ETV6 BAP and EWSR1 BAP respectively. ETV BAP FISH (a-c): a. Metaphase FISH exhibiting an intact ETV6 (yellow signal) on a normal chromosome 12 and 5′ETV6 (red signal) on an abnormal chromosome 22; b. Metaphase; c. Karyotype. EWSR1 BAP FISH (d-f): d. Metaphase FISH exhibiting an intact EWSR1 (yellow signal) on a normal chromosome 22 and 3′EWSR1 (green signal) on an abnormal chromosome 12; e. Metaphase; f. Karyotype
Due to the complexity of these chromosomal aberrations and a low resolution of the available karyogram, the derivative chromosomes were drawn by using the online CyDAS software [9], and the corresponding FISH signals were labeled (Fig. 2). Neither MYC gene rearrangement nor BCR/ABL fusion were detected in the BM specimen.
Karyograms of normal chromosomes 12 and 22, abnormal chromosomes 12 (der(12)) and 22 (der922)) drawn by using CyDAS program [9] with indication of sites and colors of ETV6 BAP and EWSR1 BAP of FISH tests in this study
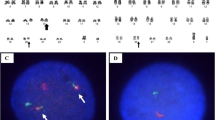
Interphase FISH performed on the formalin-fixed, paraffin-embedded LN sample showed that 90 % of cells had the same EVT6 and EWSR1 signal patterns as detected in the BM (images not shown). Therefore, the same chromosomal aberrations have been confirmed on FFPE tissues of the LN biopsy as well. We combined morphologic and FISH analysis as described previously [10, 11] to further characterize the type of BM cells carrying the chromosomal aberrations described above. As shown in Fig. 3, all cells in the BM specimen were morphologically and immunopheno typically normal (Fig. 3a, H.E. staining image), but FISH tests using ETV6 BAP probe on the same slide detected a positive signal pattern for ETV6 rearrangement and partial deletion of 3′ETV6 (Fig. 3b, FISH test image).
Combined morphology and FISH analysis on the same bone marrow slide to further characterize the type(s) of cells carrying the chromosomal aberrations described above. a H.E. staining image (100×) showing all cells morphologically normal; b FISH test using ETV6 BAP probe showing that in the same field as the H.E. staining image, majority of the cells have exhibited two-fusion/yellow signals pattern, except two maturing myelocytes (pointed with green arrows in both a and b) showing one-fusion/yellow, one-green signals pattern, indicating deletion of one copy of the 3′ETV6 (red signal)
Morphologic and flow cytometry immunophenotypic analyses
Morphological examination of hematoxylin-eosin-stained histologic sections of BM biopsy specimen and Romanowsky stained PB and/or BM aspirate smears did not show any morphologic evidence of disease. Cell markers including CD2, CD4, CD5, CD7, CD13, CD14, CD15, CD19, CD22, CD33, CD34, CD36, CD38, CD45, CD56, CD64, CD117, CD123, HLA-DR (Becton-Dickinson Biosciences, San Jose, CA) were assessed by flow cytometry immunophenotyping [6, 10, 11]. No evidence of a CD123 positive, CD4 positive cell population was detected.
Molecular testing
The BM sample was screened for somatic mutations using a clinically validated next-generation sequencing (NGS)-based 28-gene assay [6]. The genes in this panel included: ABL1, ASXL1, BRAF, DNMT3A, EGFR, EZH2, FLT3, GATA1, GATA2, HRAS, IDH1, IDH2, IKZF2, JAK2, KIT, KRAS, MDM2, MLL, MPL, MYD88, NOTCH1, NPM1, NRAS, PTPN11, RUNX1, TET2, TP53, and WT1. No mutations in any of the genes assessed were detected in the BM sample.
Discussion
We report the first case of BPDCN that carried a translocation between chromosomes 12 and 22, followed by a subsequent pericentric inversion of the abnormal chromosome 12, and that resulted in a simultaneous partial deletion of 3′ETV6 and 5′EWSR1. Based on separate locations of the remaining 5′ETV6 and 3′EWSR1 (Fig. 2), there is unlikely an ETV6/EWSR1 fusion gene present in this case. Importantly, these aberrations were detected in BPDCN cells in the LN as well as in hematopoietic precursors in a BM samples that had no evidence of involvement by BPDCN.
The cytogenetic characterization of BPDCN is not well established, mostly due to the rarity of this disease and its relatively recent recognition and diagnostic characterization. In one of the largest cohorts to date, Leroux et al. [4] reported that 14 of 21 cases of CD4 + CD56+ DC2 acute leukemia/BPDCN had an abnormal karyotype, which was further characterized using interphase FISH, metaphase FISH, whole chromosome painting (WCP) and spectral karyotyping (SKY). These analyses delineated six major chromosomal targets for this disease, including 5q (72 %), 12p (64 %), 13q (64 %), 6q (50 %), 15q (43 %) and 9 (28 %). Additional smaller studies and reports showed similar findings.
In Table 1, we have summarized a total of 46 BPDCN cases with abnormal karyotypes, 45 from previous literature reports [4, 6, 12–28] plus the case presented in this report. A minor modification has been made to some of the cases from previous literature reports in order to follow the ISCN 2013 nomenclature guidelines [8] as well as to integrate all findings derived by means other than conventional analysis (e.g., FISH and SKY) into the description of an abnormal karyotype. A similar number of BPDCN cases with possible chromosomal abnormalities are not included in this table, mainly due to a lack of complete karyotype description in these literature [29–37]. In addition, our analysis has focused on cytogenetic alterations and does not include mutations in genes such as TET2 that have been shown to be present in a sizeable subset of BPDCN [6, 38]. The mutational landscape of BPDCN is beyond the scope of this review.
Thirty-four of 46 (74 %) cases of BPDCN reported had a complex karyotype (at least 3 chromosomal aberrations including at least one structural aberration), indicating that multiple recurrent chromosomal abnormalities are very common. The frequency of involvement of each chromosome is listed in Table 2. Of interest, our literature review identified the same six major chromosomal aberrancies reported by Leroux et al. [4], but with a deviant frequency for each chromosome as follows: 6 (20/46, 43 %), 12 (20/46, 43 %), 13 (20/46, 43 %), 9 (17/46, 37 %), 15 (17/46, 37 %), and 5 (15/46, 33 %). Whereas numerical aberrations were detected frequently for chromosomes 13 (18/20, 90 %), 9 (12/17, 71 %) and 15 (9/17, 53 %), we observed structural aberrations more commonly in chromosomes 6 (20/20, 100 %), 5 (14/15, 93 %) and 12 (16/20, 80 %). Further analysis examining the breakpoints and consequences of the above mentioned structural aberrations has revealed that 5q-, 6q- and/or 12p- were common with a frequency of 93 % for 5q-, 90 % for 6q-, and 88 % for 12p-. This phenomenon has been observed by Lucioni et al. [29] in a study of 21 BPDCN cases by using an array-based comparative genomic hybridization (aCGH) assay as well as by other research groups [5, 30–34]. We have an ongoing project studying the correlation between the complexity of the karyotype and outcome of this disease in patients who had been seen and followed-up in our institute.
The 12p- is considered to be one of the most common structural aberrations in BPDCN. Two major target genes/loci located on the 12p have been identified as potential hotspots in the literature, CDKN1B and ETV6. The CDKN1B gene is located on 12p13.1p12 spanning from 12,717,270 to 12,722,383 (5114 bp, GRCh.38.p2). The protein encoded by this gene, p27 (also known as KIP1), is a kinase inhibitor and an atypical tumor suppressor through regulation of the activity of cyclin-dependent kinases (Cdks). Therefore, dysfunction of CDKN1B plays a role in pathogenesis and metastasis of multiple cancers, such as breast cancer, prostate cancer and leukemia [39]. Loss of the CDKN1B locus has been reported in over 60 % of BPDCN cases, including cases with a normal karyotype [5, 29–32]. Due to the size of this gene, its loss has been detected only by array comparative genomic hybridization assay in the literature. Immunohistochemistry studies have further confirmed weak expression of p27 protein in almost all these cases. ETV6, also known as TEL, THC5 or TEL/ABL gene, is located on 12p13 spanning from 11,649,854 to 11,895,402 (245,549 bp), less than 1 Mb apart from CDKN1B. Both genes were simultaneously lost in the cases mentioned above. ETV6 is a gene with the characteristics of a tumor suppressor gene and encodes an ETS family transcription factor. ETV6 has been shown to play an important role in the pathogenesis of various types of leukemia, mostly through forming fusion genes with over 40 different translocation partners [40, 41]. In BPDCN (Tables 1 and 2), over 40 % of cases had a structural abnormality involving 12p, very likely resulting in a 12p- at various breakpoints and including a deletion of CDKN1B and ETV6 in most cases by roughly analyzing the breakpoints. Some of these cases might have had an ETV6 rearrangement as well. ETV6 (approximately 250 Kb of size) is a good target for FISH testing and other studies have demonstrated a deletion or rearrangement of ETV6 by FISH testing in BPDCN cases with or without an obvious 12p-; the deletion can be monoallelic or biallelic [5, 29]. Therefore, 12p-, more specifically a deletion or rearrangement of CDKN1B and/or ETV6, may deserve further investigations as potential markers of BPDCN.
Chromosome 22 abnormalities appear to be rare in BPDCN (5/46, 11 %) (Tables 1 and 2). Three cases reported so far had a numerical change and 2 other cases (including the one in this study) had a structural aberration of chromosome 22. However, chromosomal aberrations involving a chromosome 22 might have been underestimated, most likely due to technical limitations including: size of chromosome 22 (one of the smallest chromosomes with limited banding patterns); extremely low karyotyping resolution in cancer cases; and high percentage of complex karyotypes and marker chromosomes in BPDCN cases. In our case, a EWSR1 rearrangement with a deletion of the 5′EWSR1 was detected. EWSR1 rearrangement has been reported mostly in soft tissue tumors [42–44], also rarely in hematologic malignancies [37, 45, 46], but only in one BPDCN case previously [37]. The EWSR1 gene has a large number of fusion partners, including members of the ets family, such as FLI1, ERG and ETV1 but not ETV6 [42–44]. As mentioned above, according to the locations of the remaining 5′ETV6 and 3′EWSR1, an ETV6/EWSR1 fusion can be excluded in this case. However, due to occurrence of inversion in the affected chromosome 12 and partial deletion of both 3′ETV6 and 5′EWSR1, gene fusions between ETV6 and a partner gene on 22q as well as between EWSR1 and a partner gene on 12q cannot be completely excluded. The biological implications of these EWSR1 aberrations are unknown.
Clinical manifestation of BPDCN vary among patients. Most BPDCN patients present with one or more skin lesions with or without BM or LN involvement. The patient in this study presented with progressive LN enlargement as the only notable finding. His BM tested by multiple means was negative for BPDCN involvement. However, the same chromosomal aberrations were detected in his LN with BPDCN as well as BM without BPDCN. Combined morphologic and FISH analysis further confirmed that the BM cells carrying the chromosomal aberrations were morphologically normal. One explanation is that the chromosomal abnormalities in our case may constitute an initiating event within a hematopoietic stem cell precursor and that a second hit in a cell capable of acquiring phenotypic features of plasmacytoid dendritic cells is required for BPDCN to develop (Fig. 3). This hypothesis is intriguing particularly since a link between BPDCN and myeloid malignancies has long been observed, even though the pathogenic link between these entities remains unknown. Indeed, many patients with BPDCN have shown to develop acute myeloid leukemia (AML) [3, 6, 28]. In addition, a sizeable subset of BPDCN involving the BM has been reported to be associated with myelodysplastic features at the morphologic and/or cytogenetic levels [6]. Another explanation is that a minimal BM dissemination of BPDCN and growth advantage of neoplastic cells in ex vivo culture may contribute to the results in our case.
In summary, over 50 % of BPDCN cases have chromosomal abnormalities, with more than 70 % of BPDCN cases exhibiting a complex karyotype. Monosomy 13/13q-, 12p-, 6q-, monosomy 15/15q-, 5q- and monosomy 9 are characteristic chromosomal abnormalities in BPDCN. The 12p- is one of the most common structural aberrations in BPDCN, and a deletion/rearrangement of CDKN1B and/or ETV6 on 12p is often detected. These two genes, together with EWSR1 on 22q, may deserve further investigations as potential BPDCN markers.
Conclusion
This is the first case of BPDCN that carried a translocation between chromosomes 12 and 22, followed by a subsequent pericentric inversion of the abnormal chromosome 12, and that resulted in a simultaneous partial deletion of 3′ETV6 and 5′EWSR1. Analyzing all 45 BPDCN cases with abnormal karyotypes available in the literature plus this case, 6 major chromosomal targets are identified in BPDCN: chromosomes 6 (20/46, 43 %), 12 (20/46, 43 %), 13 (20/46, 43 %), 9 (17/46, 37 %), 15 (17/46, 37 %), and 5 (15/46, 33 %). Deletion of 12p (12p-) is one of the most common structural aberrations, and the ETV6 and CDKN1B on 12p deserve further investigations as potential markers of BPDCN.
Consent
Written informed consent was obtained from the patient for publication of this Case report and any accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal.
Abbreviations
- aCGH:
-
array-based comparative genomic hybridization
- BM:
-
bone marrow
- BPDCN:
-
blastic plasmacytoid dendritic cell neoplasm
- Cdks:
-
cyclin-dependent kinases
- FISH:
-
fluorescence in situ hybridization
- LN:
-
lymph nodes
- PB:
-
peripheral blood
- SKY:
-
spectral karyotyping
- WCP:
-
whole chromosome painting
References
Facchetti F, Jones DM, Petrella T. Blastic plasmacytoid dendritic cell neoplasm. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al., editors. WHO Classification of Tumors of Haematopoietic and Lymphoid Tissues. IARC. Lyon: WHO Press; 2008. p. 145–7.
Brody JP, Allen S, Schulman P, Sun T, Chan WC, Friedman HD, et al. Acute agranular CD4-positive natural killer cell leukemia. Comprehensive clinicopathologic studies including virologic and in vitro culture with inducing agents. Cancer. 1995;75(10):2474–83.
Khoury JD, Medeiros LJ, Manning JT, Sulak LE, Bueso-Ramos C, Jones D. CD56(+) TdT(+) blastic natural killer cell tumor of the skin: a primitive systemic malignancy related to myelomonocytic leukemia. Cancer. 2002;94(9):2401–8. doi:10.1002/cncr.10489.
Leroux D, Mugneret F, Callanan M, Radford-Weiss I, Dastugue N, Feuillard J, et al. CD4(+), CD56(+) DC2 acute leukemia is characterized by recurrent clonal chromosomal changes affecting 6 major targets: a study of 21 cases by the Groupe Francais de Cytogenetique Hematologique. Blood. 2002;99(11):4154–9.
Petrella T, Bagot M, Willemze R, Beylot-Barry M, Vergier B, Delaunay M, et al. Blastic NK-cell lymphomas (agranular CD4 + CD56+ hematodermic neoplasms): a review. Am J Clin Pathol. 2005;123(5):662–75.
Alayed K, Patel KP, Konoplev S, Singh RR, Routbort MJ, Reddy N, et al. TET2 mutations, myelodysplastic features, and a distinct immunoprofile characterize blastic plasmacytoid dendritic cell neoplasm in the bone marrow. Am J Hematol. 2013;88(12):1055–61. doi:10.1002/ajh.23567.
Khoury JD, Sen F, Abruzzo LV, Hayes K, Glassman A, Medeiros LJ. Cytogenetic findings in blastoid mantle cell lymphoma. Hum Pathol. 2003;34(10):1022–9.
ISCN. An International System for Human Cytogenetic Nomenclature (2013). Basel, Switzerland: S. Kager AG; 2013. p. 2013.
Hiller B, Bradtke J, Balz H, Rieder H. CyDAS: a cytogenetic data analysis system. Bioinformatics. 2005;21(7):1282–3. doi:10.1093/bioinformatics/bti146.
Wang SA, Hutchinson L, Tang G, Chen SS, Miron PM, Huh YO, et al. Systemic mastocytosis with associated clonal hematological non-mast cell lineage disease: clinical significance and comparison of chomosomal abnormalities in SM and AHNMD components. Am J Hematol. 2013;88(3):219–24. doi:10.1002/ajh.23380.
Tang G, Goswami RS, Liang CS, Bueso-Ramos CE, Hu S, DiNardo C, et al. Isolated del(5q) in Patients Following Therapies for Various Malignancies May Not All Be Clinically Significant. Am J Clin Pathol. 2015;144(1):78–86. doi:10.1309/AJCPBADO22WXOFHJ.
Petrella T, Dalac S, Maynadie M, Mugneret F, Thomine E, Courville P, et al. CD4+ CD56+ cutaneous neoplasms: a distinct hematological entity? Groupe Francais d’Etude des Lymphomes Cutanes (GFELC). Am J Surg Pathol. 1999;23(2):137–46.
Bayerl MG, Rakozy CK, Mohamed AN, Vo TD, Long M, Eilender D, et al. Blastic natural killer cell lymphoma/leukemia: a report of seven cases. Am J Clin Pathol. 2002;117(1):41–50. doi:10.1309/U72E-XD57-NF4J-XVG8.
Rakozy CK, Mohamed AN, Vo TD, Khatib G, Long PM, Eilender D, et al. CD56+/CD4+ lymphomas and leukemias are morphologically, immunophenotypically, cytogenetically, and clinically diverse. Am J Clin Pathol. 2001;116(2):168–76. doi:10.1309/FMYW-UL3G-1D7J-ECQF.
Zhang K, Prichard JW, Brown RE. Blastic natural killer (NK) cell leukemia (agranular CD4 + CD56+ leukemia). Ann Clin Lab Sci. 2006;36(2):212–5.
Wilson CS, Medeiros LJ. Extramedullary Manifestations of Myeloid Neoplasms. Am J Clin Pathol. 2015;144(2):219–39. doi:10.1309/AJCPO58YWIBUBESX.
Patel JL, Shetty S, Salama ME. An unusual case of cutaneous blastic plasmacytoid dendritic cell neoplasm with concomitant B-cell lymphoproliferative disorder. Am J Dermatopathol. 2011;33(3):e31–6. doi:10.1097/DAD.0b013e3181de9ce0.
Goren Sahin D, Akay OM, Uskudar Teke H, Andic N, Gunduz E, Gulbas Z. Blastic plasmacytoid dendritic cell leukemia successfully treated by autologous hematopoietic stem cell transplantation to a remission of 48-month duration. Case Rep Hematol. 2013;2013:471628. doi:10.1155/2013/471628.
Anargyrou K, Paterakis G, Boutsis D, Politou M, Papadhimitriou SI, Siakandaris M, et al. An unusual case of CD4+ CD7+ CD56+ acute leukemia with overlapping features of type 2 dendritic cell (DC2) and myeloid/NK cell precursor acute leukemia. Eur J Haematol. 2003;71(4):294–8.
Karube K, Ohshima K, Tsuchiya T, Yamaguchi T, Suefuji H, Suzumiya J, et al. Non-B, non-T neoplasms with lymphoblast morphology: further clarification and classification. Am J Surg Pathol. 2003;27(10):1366–74.
Rossi JG, Felice MS, Bernasconi AR, Ribas AE, Gallego MS, Somardzic AE, et al. Acute leukemia of dendritic cell lineage in childhood: incidence, biological characteristics and outcome. Leuk Lymphoma. 2006;47(4):715–25. doi:10.1080/10428190500353216.
Eguaras AV, Lo RW, Veloso JD, Tan VG, Enriquez ML, Del Rosario ML. CD4+/CD56+ hematodermic neoplasm: blastic NK cell lymphoma in a 6-year-old child: report of a case and review of literature. J Pediatr Hematol Oncol. 2007;29(11):766–9. doi:10.1097/MPH.0b013e318159a4e6.
Chang HJ, Lee MD, Yi HG, Lim JH, Lee MH, Shin JH, et al. A case of blastic plasmacytoid dendritic cell neoplasm initially mimicking cutaneous lupus erythematosus. Cancer Res Treat. 2010;42(4):239–43. doi:10.4143/crt.2010.42.4.239.
Nakamura Y, Kayano H, Kakegawa E, Miyazaki H, Nagai T, Uchida Y, et al. Identification of SUPT3H as a novel 8q24/MYC partner in blastic plasmacytoid dendritic cell neoplasm with t(6;8)(p21;q24) translocation. Blood Cancer J. 2015;5, e301. doi:10.1038/bcj.2015.26.
Agapidou A, Vakalopoulou S, Markala D, Chadjiaggelidou C, Tzimou M, Papadopoulou T, et al. Blastic plasmacytoid dendritic cell neoplasm: single-center experience with two cases in one year. Turk J Haematol. 2014;31(4):422–3. doi:10.4274/tjh.2013.0404.
DiGiuseppe JA, Louie DC, Williams JE, Miller DT, Griffin CA, Mann RB, et al. Blastic natural killer cell leukemia/lymphoma: a clinicopathologic study. Am J Surg Pathol. 1997;21(10):1223–30.
Kameoka J, Ichinohasama R, Tanaka M, Miura I, Tomiya Y, Takahashi S, et al. A cutaneous agranular CD2–CD4+ CD56+ “lymphoma”: report of two cases and review of the literature. Am J Clin Pathol. 1998;110(4):478–88.
Yu S, Kwon MJ, Kim K, Koo DH, Woo HY, Park H. A rare case of acute leukemic presentation of blastic plasmacytoid dendritic cell neoplasm without cutaneous lesions. Ann Lab Med. 2014;34(2):148–51. doi:10.3343/alm.2014.34.2.148.
Lucioni M, Novara F, Fiandrino G, Riboni R, Fanoni D, Arra M, et al. Twenty-one cases of blastic plasmacytoid dendritic cell neoplasm: focus on biallelic locus 9p21.3 deletion. Blood. 2011;118(17):4591–4. doi:10.1182/blood-2011-03-337501.
Pagano L, Valentini CG, Pulsoni A, Fisogni S, Carluccio P, Mannelli F, et al. Blastic plasmacytoid dendritic cell neoplasm with leukemic presentation: an Italian multicenter study. Haematologica. 2013;98(2):239–46. doi:10.3324/haematol.2012.072645.
Reineks EZ, Osei ES, Rosenberg A, Auletta J, Meyerson HJ. CD22 expression on blastic plasmacytoid dendritic cell neoplasms and reactivity of anti-CD22 antibodies to peripheral blood dendritic cells. Cytometry B Clin Cytom. 2009;76(4):237–48. doi:10.1002/cyto.b.20469.
Wiesner T, Obenauf AC, Cota C, Fried I, Speicher MR, Cerroni L. Alterations of the cell-cycle inhibitors p27(KIP1) and p16(INK4a) are frequent in blastic plasmacytoid dendritic cell neoplasms. J Invest Dermatol. 2010;130(4):1152–7. doi:10.1038/jid.2009.369.
Dharmani PA, Mittal NM, Subramanian PG, Galani K, Badrinath Y, Amare P, et al. Blastic plasmacytoid dendritic cell neoplasm: report of two pediatric cases. Indian J Pathol Microbiol. 2015;58(1):72–6. doi:10.4103/0377-4929.151193.
Roos-Weil D, Dietrich S, Boumendil A, Polge E, Bron D, Carreras E, et al. Stem cell transplantation can provide durable disease control in blastic plasmacytoid dendritic cell neoplasm: a retrospective study from the European Group for Blood and Marrow Transplantation. Blood. 2013;121(3):440–6. doi:10.1182/blood-2012-08-448613.
Shaw PH, Cohn SL, Morgan ER, Kovarik P, Haut PR, Kletzel M, et al. Natural killer cell lymphoma: report of two pediatric cases, therapeutic options, and review of the literature. Cancer. 2001;91(4):642–6.
Gao NA, Wang XX, Sun JR, Yu WZ, Guo NJ. Blastic plasmacytoid dendritic cell neoplasm with leukemic manifestation and ETV6 gene rearrangement: A case report. Exp Ther Med. 2015;9(4):1109–12. doi:10.3892/etm.2015.2236.
Cao Q, Liu F, Niu G, Xue L, Han A. Blastic plasmacytoid dendritic cell neoplasm with EWSR1 gene rearrangement. J Clin Pathol. 2014;67(1):90–2. doi:10.1136/jclinpath-2013-201792.
Menezes J, Acquadro F, Wiseman M, Gómez-López G, Salgado R, Talavera-Casañas J, et al. Exome sequencing reveals novel and recurrent mutations with clinical impact in blastic plasmacytoid dendritic cell neoplasm. Leukemia. 2014;28(4):7.
Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008;8(4):253–67. doi:10.1038/nrc2347.
Bohlander SK. ETV6: a versatile player in leukemogenesis. Semin Cancer Biol. 2005;15(3):162–74. doi:10.1016/j.semcancer.2005.01.008.
De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Basinko A, De Braekeleer M. ETV6 fusion genes in hematological malignancies: a review. Leuk Res. 2012;36(8):945–61. doi:10.1016/j.leukres.2012.04.010.
Khoury JD. Ewing sarcoma family of tumors. Adv Anat Pathol. 2005;12(4):212–20.
Khoury JD. Ewing sarcoma family of tumors: a model for the new era of integrated laboratory diagnostics. Expert Rev Mol Diagn. 2008;8(1):97–105. doi:10.1586/14737159.8.1.97.
Cantile M, Marra L, Franco R, Ascierto P, Liguori G, De Chiara A, et al. Molecular detection and targeting of EWSR1 fusion transcripts in soft tissue tumors. Med Oncol. 2013;30(1):412. doi:10.1007/s12032-012-0412-8.
Jakovljevic G, Nakic M, Rogosic S, Kardum-Skelin I, Mrsic-Davidovic S, Zadro R, et al. Pre-B-cell acute lymphoblastic leukemia with bulk extramedullary disease and chromosome 22 (EWSR1) rearrangement masquerading as Ewing sarcoma. Pediatr Blood Cancer. 2010;54(4):606–9. doi:10.1002/pbc.22365.
Haresh KP, Joshi N, Gupta C, Prabhakar R, Sharma DN, Julka PK, et al. Granulocytic sarcoma masquerading as Ewing’s sarcoma: a diagnostic dilemma. J Cancer Res Ther. 2008;4(3):137–9.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
All authors declare that they have no competing interests.
Authors’ contributions
All authors have participated in the clinical care and clinical diagnosis of this patient, writing and editing of the manuscript. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Tang, Z., Tang, G., Wang, S.A. et al. Simultaneous deletion of 3′ETV6 and 5′EWSR1 genes in blastic plasmacytoid dendritic cell neoplasm: case report and literature review. Mol Cytogenet 9, 23 (2016). https://doi.org/10.1186/s13039-016-0232-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-016-0232-1