Abstract
Background
Sugarcane mosaic virus (SCMV) is the prevalent virus inducing maize dwarf mosaic and sugarcane mosaic diseases in China. According to the phylogenetic results of the complete genomic and coat protein gene sequences, SCMV was divided into four or five molecular groups, respectively. Previously, we detected SCMV isolates of group SO from Canna spp. in Ji’nan, Shandong province, China.
Findings
In this study, we collected two SCMV isolates infecting Canna spp. in Ji’nan (Canna-Ji’nan) and Tai’an (Canna-Tai’an) of Shandong, China. Their complete genome sequences had genome of 9576 nucleotides and contained a large open reading frame encoding a polyprotein of 3063 amino acids. The phylogenetic analysis showed that the both Canna-Ji’nan and Canna-Tai’an were clustered into an independent group based on the complete genome sequence.
Conclusion
In this study, we report the complete genome sequences of SCMV infecting Canna spp. from Ji’nan and Tai’an. This is the first report on SCMV belonging to SO group.
Similar content being viewed by others
Body of text
Sugarcane mosaic virus (SCMV) is a species of the genus Potyvirus in the Potyviridae family and mainly infects plants of monocotyledons including maize, sorghum, sugarcane and canna. It is the major pathogen of sugarcane mosaic disease and maize dwarf mosaic disease and causes significant yield losses in China [1, 2]. SCMV has flexuous filamental particles of 700~ 750 nm long and can be transmitted by aphids in a non-persistent manner. The SCMV genome is single-stranded, positive RNA containing a single open reading frame (ORF), which code a large polyprotein and a truncated frame-shift product [3]. SCMV isolates can be clustered to four groups according to the phylogenetic analysis of the complete genomic sequences [4] . However, according to the phylogenetic results of coat protein genes, SCMV isolates are divided into five groups (A to E) specific to host origins [4]. Isolates of group E (SO) have been reported from sugarcane (Saccharum officinarum) in Vietnam and China [4] and are therefore designated as group SO. In 2015 and 2016, SCMV isolates of group SO was reported to infect Cannas spp. [5]. But so far there is no information on the complete genomic sequences of SCMV from group SO available.
Provenance of virus material
Plant of Canna spp. showing symptoms of severe mosaic and dwarf were observed at incidence of 20%~ 30%. Two symptomatic leaf samples were collected in 2015 and 2016, respectively, from the parks of Ji’nan (N36°43′, E117°36′) and Tai’an (N36°10′, E117°10′), Shandong Province, China. Total RNA was extracted using the RNAprep Pure Plant Kit (Polysaccharides & Polyphenolics-rich) (Tiangen, Beijing, China). The first-strand cDNA was synthesized using Moloney MLV reverse transcriptase (Takara, Dalian, China) with primer 5’-GGTCGACTGCAGGATCCAAGCT15–3′ [6]. Degenerate primers was designed according to the conserved regions of potyviruses and were used to amplify different genomic parts of virus via RT-PCR [7]. To determine the genomic ends, rapid amplification of cDNA ends (RACE) was conducted using the 5’-Full RACE Kit with TAP (Takara, Dalian, China) with specific primers. An additional WORD file shows the primer sequences in more detail (see Additional file 1). DNASTAR 7.1.0 package (DNASTAR Inc., Madison, WI, USA) was used to assemble the sequences. Phylogenetic relationships were inferred by the Neighbor-Joining and Maximum Likelihood methods with 1000 bootstrap replicates using MEGA6.06.
Sequence properties
Both of the two Canna samples were positive to SCMV antiserum in ELISA detection. The SCMV infecting Canna plants in Ji’nan and Tai’an were designated as Canna-Ji’nan and Canna-Tai’an accordingly. The complete genome of SCMV Canna-Ji’nan and Canna-Tai’an have been deposited in the GenBank database and allocated the accession numbers KY548506 and KY548507, respectively. The complete genome of both SCMV canna isolates were 9576 nucleotides (nt) in length excluding the poly (A) tails. Their 5’-UTRs and 3’-UTRs were 148 nt and 238 nt, respectively. The conserved potyboxes ‘a’ and ‘b’ in 5’-UTR [8] were identified as A19CAACAC25 and C38CAAGCA44. The first in-frame AUG149–151 situated in the context CGAGAUGGC was presumed to be the initiation codon of the polyprotein translation. The ORF of SCMV canna isolate was 9192 nt, encoding a polyprotein 3063 amino acids (aa). The polyprotein of SCMV is processed into ten mature functional proteins, and the nine putative protease cleavage sites were identified (Table 1) [9]. Unlike Y/A in other SCMV isolates, the cleavage site at the P1/HC-Pro junction of Canna-Ji’nan and Canna-Tai’an was Y/S, which was also found in that of Johnsongrass mosaic virus (JGMV). The cleavage site at VPg/NIa-Pro junction of other SCMV isolates was E/S, while that of Canna-Ji’nan and Canna-Tai’an was E/A. E/A was also found in the cleavage site at VPg/NIa-Pro junction of Tobacco vein banding mosaic virus (TVBMV) [6]. The highly conserved G1~2A6~ 7 (G2 686GAAAAAA2 693) motif was also identified within the P3 protein, producing PIPO with the + 2 reading frameshift.
The complete genome of SCMV Canna-Ji’nan and Canna-Tai’an shared nt identity of 98.0% and aa identity of 98.8%. At the complete genome level, Canna-Ji’nan shared the highest nt identities of 76.5% with isolates Henan (AF495510) and SX (AY569692), aa identities of 85.5% with isolates Xiangshan (AJ310103) and Yuhang (AJ310104) from China and isolate sp. (AM110759) from Spain; while Canna-Tai’an shared the highest nt identities of 76.6% with isolates Beijing (AY042184) from China and JAL-1 (GU474635) from Mexcio, aa identities of 85.6% with isolates Xiangshan (AJ310103) and Yuhang (AJ310104). At the coat protein gene level, Canna-Ji’nan and Canna-Tai’an shared nt identities of 87–91% and 87–92% with isolates from Yunnan province, China and Vietnam (FM998893-FM998896 and DQ925427-DQ925428) in GenBank, which clustered into SO group based on cp gene. The most variable gene P1 shared the lowest nt identities of 56.7–62.2% and aa identities of 51.5–55.8%, while PIPO shared the highest nt identities of 87.1–91.2% and aa identities of 85.0–92.5% among the eleven mature proteins between SCMV canna isolates and other isolates. An additional WORD file shows the nt and aa identities in more detail (see Additional file 2).
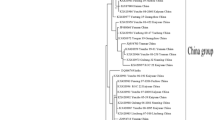
A phylogenetic tree was constructed with the complete genomic sequences of Canna-Jinan, Canna-Tai’an and other 29 SCMV isolates, using that of a MDMV isolate (AJ001691) as the out-group. The results showed that these 31 SCMV isolates were clustered to five groups. Group I contained 12 SCMV isolates from maize and was divided into two subgroups. The isolates of subgroup IA were collected from Europe, Africa and America, while those of subgroup IB were from China. Group II contained 11 isolates and was also divided into two subgroups. Subgroup IIA contained four isolates from maize, three from Africa and one from America; subgroup IIB contained six isolates from sugarcane in America and Asia, the other one from maize in Iran was actually a recombinant [10]. Group III contained four isolates from China, among which three from sugarcane and one from maize. Group IV included only two isolates from maize. BD8 was a high virulent isolate in China and had spread rapidly [4, 11]; MO1 was a isolate from Ecuador. Group V(SO) contained the Canna-Ji’nan and Canna-Tai’an. Because there was no complete genomic sequence of SO group available, SCMV was clustered into four groups in phylogenetic trees in the previous studies [4] (Fig. 1).
Phylogenetic tree of SCMV constructed with the complete genome sequences using the neighbor-joining method in MEGA6.06 program. The SCMV isolates were listed as accession number/isolate name/host plant/country of origin. Bootstrap analysis was applied using 1000 replicates. Only bootstrap values higher than 70% were shown. One Maize dwarf mosaic virus isolate (AJ001691) was used as an out-group. ◆ indicated the isolate in this study. The scale bars showed a genetic distance of 0.05
Canna is commonly cultivated as an important ornamental plant in the world. The canna virus disease has been increasing in the past years. The viruses reported to infect canna included Cucumber mosaic virus (CMV) [12, 13], Tomato aspermy virus (TAV) [14], Bean yellow mosaic virus (BYMV) [15], Canna yellow mottle virus (CaYMV) [12], Canna yellow streak virus (CaYSV) [16] and Sugarcane mosaic virus (SCMV) [5]. We hypothesized that the SCMV isolates of SO lineage were transmitted to Canna plants by aphids and experienced distinct evolution events. To our knowledge, this is first complete genomic sequences of SCMV SO group and also the first complete genomic sequences of SCMV infecting canna.
Abbreviations
- aa:
-
amino acids
- BYMV:
-
Bean yellow mosaic virus
- CaYMV:
-
Canna yellow mottle virus
- CaYSV:
-
Canna yellow streak virus
- CMV:
-
Cucumber mosaic virus
- JGMV:
-
Johnsongrass mosaic virus
- kDa:
-
kilodalton
- MDMV:
-
Maize dwarf mosaic virus
- nm:
-
nanometer
- nt:
-
nucleotides
- ORF:
-
open reading frame
- RdRp:
-
RNA-dependent RNA polymerase
- SCMV:
-
Sugarcane mosaic virus
- TAV:
-
Tomato aspermy virus
- TVBMV:
-
Tobacco vein banding mosaic virus
- UTR:
-
untranslated region
References
Agnihotri VP. Current sugarcane disease scenario and management strategies. Indian Phytopathology. 1996;49:109–26.
Ricaud C, Egan BT, Gillaspie AG, Hughes CG: Diseases of sugarcane: major diseases. Elsevier B.V.; 1989.
Chung BYW, Miller WA, Atkins JF, Firth AE. An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci U S A. 2008;105:5897–902.
Gao B, Cui XW, Li XD, Zhang CQ, Miao HQ. Complete genomic sequence analysis of a highly virulent isolate revealed a novel strain of Sugarcane mosaic virus. Virus Genes. 2011;43:390–7.
Tang W, Xu XH, Sun HW, Li F, Gao R, Yang SK, Lu XB, Li XD. First Report of Sugarcane mosaic virus infecting Canna spp in China. Plant Disease. 2016;100:2541.
Yu XQ, Lan YF, Wang HY, Liu JL, Zhu XP, Valkonen JPT, Li XD. The complete genomic sequence of Tobacco vein banding mosaic virus and its similarities with other potyviruses. Virus Genes. 2007;35:801–6.
Chen J, Chen J, Adams MJ. Characterisation of potyviruses from sugarcane and maize in China. Arch Virol. 2002;147:1237–46.
Turpen T. Molecular cloning of a potato virus Y genome: nucleotide sequence homology in non-coding regions of potyviruses. J Gen Virol. 1989;70:1951–60.
Adams MJ, Antoniw JF, Beaudoin F. Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol Plant Pathol. 2005;6:471–87.
Moradi Z, Mehrvar M, Nazifi E, Zakiaghl M. The complete genome sequences of two naturally occurring recombinant isolates of Sugarcane mosaic virus from Iran. Virus Genes. 2016;52:270–80.
Yan ZY, Cheng DJ, Liu J, Tian YP, Zhang SB, Li XD. First report of Sugarcane mosaic virus group IV isolates from the corn production fields in China. Plant Dis. 2016;100:1508.
Lockhart BEL. Occurrence of Canna yellow mottle virus in North America. ISHS Acta Horticulturae. 1988;234:69–72.
Fisher JR, Sanchez-Cuevas MC, Nameth ST, Woods VL, Ellett CW. First report of cucumber mosaic virus in Eryngium amethystinum, Canna spp., and Aquilegia hybrids in Ohio. Plant Dis. 1997;81:1331.
Hollings M, Stone OM: Tomato aspermy virus. CMI/AAB Descriptions of Plant Viruses 1971, No. 79 Wallingford.
Skelton A, Daly M, Nixon T, Harju V, Mumford RA. First record of Bean yellow mosaic virus infecting a member of the orchid genus Dactylorhiza. Plant Pathol. 2007;56:344.
Monger WA, Harju V, Skelton A, Seal SE, Mumford RA. Canna yellow streak virus: a new potyvirus associated with severe streaking symptoms in canna. Arch Virol. 2007;152:1527–30.
Funding
This work was supported by funds from National Key Research and Development Program (2018YFD0200604), Shandong Modern Agricultural Technology & Industry System (SDAIT-02-10), Shandong ‘Double Top’ Program (SYL2017XTTD11) and ‘Taishan Scholar’ Construction Project (TS201712023).
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Author information
Authors and Affiliations
Contributions
WT, ZY, XL collected the samples, conceived and designed the experiments. WT, ZY and XX performed the experiments. WT, TZ and YT analyzed the data. XL and WT wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
SCMV primer sequences. (DOC 16 kb)
Additional file 2:
SCMV nucleotide and amino acid sequence identities. (DOC 102 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Tang, W., Yan, ZY., Zhu, TS. et al. The complete genomic sequence of Sugarcane mosaic virus from Canna spp. in China. Virol J 15, 147 (2018). https://doi.org/10.1186/s12985-018-1058-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-018-1058-8