Abstract
Background
Selenium (Se) is an essential trace element in living systems. Microorganisms play a pivotal role in the selenium cycle both in life and in environment. Different bacterial strains are able to reduce Se(IV) (selenite) and (or) Se(VI) (selenate) to less toxic Se(0) with the formation of Se nanoparticles (SeNPs). The biogenic SeNPs have exhibited promising application prospects in medicine, biosensors and environmental remediation. These microorganisms might be explored as potential biofactories for synthesis of metal(loid) nanoparticles.
Results
A strictly aerobic, branched actinomycete strain, ES2-5, was isolated from a selenium mining soil in southwest China, identified as Streptomyces sp. based on 16S rRNA gene sequence, physiologic and morphologic characteristics. Both SEM and TEM-EDX analysis showed that Se(IV) was reduced to Se(0) with the formation of SeNPs as a linear chain in the cytoplasm. The sizes of the SeNPs were in the range of 50–500 nm. The cellular concentration of glutathione per biomass decreased along with Se(IV) reduction, and no SeNPs were observed in different sub-cellular fractions in presence of NADPH or NADH as an electron donor, indicating glutathione is most possibly involved in vivo Se(IV) reduction. Strain ES2-5 was resistant to some heavy metal(loid)s such as Se(IV), Cr(VI) and Zn(II) with minimal inhibitory concentration of 50, 80 and 1.5 mM, respectively.
Conclusions
The reducing mechanism of Se(IV) to elemental SeNPs under aerobic condition was investigated in a filamentous strain of Streptomyces. Se(IV) reduction is mediated by glutathione and then SeNPs synthesis happens inside of the cells. The SeNPs are released via hypha lysis or fragmentation. It would be very useful in Se bioremediation if Streptomyces sp. ES2-5 is applied to the contaminated site because of its ability of spore reproduction, Se(IV) reduction, and adaptation in soil.
Similar content being viewed by others
Background
Selenium (Se) is an essential trace element for the adequate and healthy life of human, animal, bacterium and other living systems and has an uneven distribution in the Earth’s crust [1]. Today, selenium is well recognized to play fundamental roles on several physiological functions in diverse organisms, such as biosynthesis of selenocysteine (Sec), the 21st amino acid with specific UGA stop codon, and many selenoenzymes including formate dehydrogenase, thioredoxin reductase, and glutathione peroxidase [2–4]. In human, either Se excess or deficiency results in more than 20 kinds of symptoms such as growth retardation, endemic diseases, impaired bone metabolism and risk of diabetes [5]. Events of selenium toxicity in human have been reported in Enshi, Hubei province of China and in Indian Punjab [6]. Therefore, selenium contamination requires bioremediation initiatives especially in those geographic locations. Phylogenetically diverse microorganisms are involved in the transformation of selenium from one oxidation state to another and thus play a pivotal role on the selenium biogeochemical cycle [4, 7, 8]. Numerous bacteria are able to reduce the toxically soluble forms of Se(VI)/Se(IV) to less-toxic insoluble Se(0), visible as red-colored nanoparticles (SeNPs) [4, 9–14]. The biosynthesized SeNPs have been found applications in various fields including medicine as antimicrobial, antioxidant and anticancer agents [15–18], biosensors [19, 20] and environmental remediation [21–23].
Se(IV)-reducing bacteria generate SeNPs under aerobic and anaerobic conditions. Anaerobic Se(IV)-reducing bacteria encompassed many species such as Thauera selenatis [24], Aeromonas salmonicida [25], purple non-sulfur bacteria [11] and Shewanella oneidensis MR-1 [26]. Aerobic Se(IV)-reducing bacteria included diverse species such as Rhizobium sp. B1 [12], Stenotrophomonas maltophilia SeITE02 [27], Pseudomonas seleniipraecipitans CA5 [28], Duganella sp. and Agrobacterium sp. [13], Comamonas testosteroni S44 [29] and Bacillus mycoides [30]. Therefore, the most Se(IV)-reducing bacteria were distributed in alpha-, beta-, gamma-, delta-proteobacteria and Firmicutes.
Selenium nanoparticles were formed not only under aerobic and anaerobic conditions, but also appeared in the cytoplasm, periplasm and/or outside the cells in different bacteria [4, 9, 10, 13, 14, 24, 29, 31], implying the various mechanisms of Se(IV)-reduction in diverse microbes. One of mechanisms linking redox precipitation of both elemental sulfur and elemental selenium was observed outside sulfate-reducing bacterial cells [32]. The intracellular Se(IV) reduction was usually driven by reduced thiols such as glutathione (GSH) via the Painter reaction in Rhodospirillum rubrum, Escherichia coli and Bacillus mycoides [28, 30, 33, 34]. Moreover, diverse enzymes were responsible for Se(IV) reduction to SeNPs. The periplasmic nitrite reductase was involved in Se(IV) reduction in T. selenatis [24] and Rhizobium selenitireducens [31], while fumarate reductase catalyzed Se(IV) reduction in Shewanella oneidensis [26]. In addition, glutathione reductase and thioredoxin reductase in Pseudomonas seleniipraecipitans [28], arsenate reductase in Bacillus selenitireducens [35] and hydrogenase in Clostridium pasteurianum [36] were potentially involved in Se(IV) reduction. However, so far no gene product or enzyme solely responsible for Se(IV) reduction in aerobic bacteria has been identified in vivo.
In addition, the efflux system by which Se(0) or SeNPs deposits were exported from inside the cells to the extracellular environment still remains unknown. It was suggested that SeNPs were released into the medium via a rapid expulsion process [21] or elemental Se(0) was transported out of the cell where the SeNPs were formed [29]. The large sizes of SeNPs were also possibly released by cell lysis [37] or vesicular expulsion [9].
In this study, we isolated a filamentous actinobacterium ES2-5 from a selenium mining soil in Enshi, Hubei province of China. The process of Se(IV) reduction leading to biosynthesized SeNPs under aerobic condition was investigated using scanning electron microscopy (SEM), transmission electron microscopy (TEM) and electron dispersion spectroscopy (EDX). Evidences were provided for the SeNPs formation to be mainly in the cytoplasm of cells and then released through hyphal lysis or fragmentation. The possible mechanism of Se(IV) reduction was also proposed.
Results
Characteristics and taxonomic identification of the strain ES2-5
Strain ES2-5 was isolated from a selenium mine soil in Hubei province, China. The acidic soil (pH 4.7) had 38 mg kg−1 of total Se content and 119 mg kg−1 of total Cr content. Accordingly, the resistance of strain ES2-5 to Se(IV), Cr(VI) and other heavy metals was determined in 1/10 TSA plates. The minimal inhibitory concentrations (MIC) of Se(IV) and Cr(VI) were 50 and 80 mM, respectively. In contrast, the MICs of Zn(II) (1.5 mM), Cu(II) (0.2 mM), As(III) (0.05 mM) and Sb(III) (0.08 mM) were lower than that of Se(IV) and Cr(VI). In addition, strain ES2-5 has the ability to produce lecithinase and H2S when it grew in TSB. It was positive for motility and hemolytic reaction, whereas it was negative for utilization of citrate and hydrolysis of gelation and tyrosine.
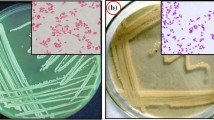
The 16S rRNA gene sequence of strain ES2-5 (1487 bp) revealed highest similarities to that of Streptomyces siamensis KC-038T (98.77 %), S. kanamyceticus NBRC 13414T (98.63 %), S. olivochromogenes NBRC 3178T (98.63 %), S. aureus NBRC 100912T (98.56 %), S. spirocerticilatus NBRC 12821T (98.43 %) and S. albiflavescens n20T (98.39 %). Phylogenetic analyze using the neighbor-joining method showed that strain ES2-5 fell in the same cluster with S. siamensis KC-038T (AB773848) and S. albiflavescens n20T (KC771426C) (Fig. 1). Moreover, strain ES2-5 formed grey, floury colonies on 1/10 TSA plates (Fig. 2a), with well growing substrate mycelia, aerial hypha and sporophores. Consequently, strain ES2-5 was characterized as Streptomyces sp. based on the phylogenetic, morphologic and some physiologic characteristics.
Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences using MEGA software version 5, showing the phylogenetic relationship of strain ES2-5 and related type strains. Bootstrap values >50 % based on 1000 replications are shown at branch nodes. Bar, 0.002 substitutions per nucleotide position
Streptomyces sp. ES2-5 reduced selenite to red elemental SeNPs. Growth of Streptomyces sp. ES2-5 on 1/10 TSA plates with 10.0 mM sodium selenite (b). LM image of SeNPs (d) on 1/10 TSA plates with 10.0 mM sodium selenite. SEM image of SeNPs (f) in 1/10 TSB broth amended with 1.0 mM sodium selenite. The a, c and e are control, respectively
Filamentous Streptomyces sp. ES2-5 was able to reduce Se(IV) to SeNPs under aerobic condition
Streptomyces sp. ES2-5 was not able to grow under anaerobic condition, indicating it is an obligate aerobe. Streptomyces sp. ES2-5 formed reddish colonies after 7 day’s incubation on 1/10 TSA plates amended with 10.0 mM selenite (Fig. 2b). The stained mycelia were observed in situ by light microscopy after 3 day’s incubation, the red-colored selenium particles were scattered away from mycelia or distributed as bean chains attaching on the mycelial surface (Fig. 2d). After 3 day’s incubation in 1/10 TSB broth, the mycelia were harvested and observed by SEM. Surprisingly, the selenium particles did not attach on the surface of mycelia but located in the mycelia as mature beans in pods (Fig. 2f). TEM of ultra-thin sections also revealed the common presence of intracellular Se(0) particles when mycelia were grown on Se(IV) (Fig. 3b–d). It was clear that the sizes of intracellular SeNPs varied from 50 to 500 nm and small SeNPs may aggregate into bigger particles. Dark, fine-grained nanoparticles were observed by EDX spectra which indicated that these nanoparticles were composed entirely of selenium as the expected emission peaks for selenium at 1.37 (Fig. 3e, f), 11.22 and 12.49 keV (data not shown) corresponding to the SeLα, SeKα, and SeKβ transitions, respectively, but EDX peaks for C, K, O, P, Cl and Ca were also produced, suggesting that these elements were in cytoplasm of cells.
The growth and capability of Streptomyces sp. ES2-5 to transform selenite to elemental selenium were tested in 1/10 TSB broth with the addition of 1.0 mM selenite (Fig. 4). On selenite-exposed cultures the growth was delayed with respect to controls, but after 24 h the biomass decreased gradually in a similar way. The formation of red cell suspension of elemental selenium started after 16 h of exposure to selenite. Streptomyces sp. ES2-5 was unable to reduce Se(IV) to elemental selenium completely. It was only able to reduce 1.0 mM Se(IV) to 0.5 mM slowly and smoothly during 52 h incubation in 1/10 TSB broth under aerobic condition.
Mechanism of Se(IV) reduction to Se(0) nanoparticles by Filamentous Streptomyces sp. ES2-5
To help understand how Se(IV) is reduced, the ability of vitro Se(IV) reduction by cultural supernatant and different cellular fractions was determined. When cultural supernatant without cells was mixed with Se(IV), the reduction of Se(IV) to red-colored precipitation and decrease of Se(IV) concentrations were not observed, indicating the reduction of Se(IV) is processed in cells. Moreover, neither red-colored precipitation nor decrease of Se(IV) concentrations appeared in the cytoplasmic fraction or in cell membrane fraction with NADPH or NADH as electron donors. These results suggest that NADPH or NADH dependent reductase and reduced chemicals are not involved in vitro Se(IV) reduction. Consequently, the concentrations of glutathione (GSH) per biomass in cells (intracellular) and in cultural broth (extracellular) were determined when Streptomyces sp. ES2-5 grew in 1/10 TSB broth at 1.0 mM concentration of selenite.
In selenite-exposed cultures the intracellular GSH content per biomass was lower than in controls during first 24 h of incubation (Fig. 5a). While, the extracellular GSH content showed an opposite pattern (Fig. 5b). After 24 h the intercellular and extracellular GSH contents in selenite-exposed cultures were similar to controls.
Discussion
Although the reduction of Se oxyanions to Se(0) nanoparticles by microorganisms has been known for some time [4, 9, 10, 13, 14, 24, 29, 38, 39], the SeNP-synthetic process and Se(IV)-reducing mechanism of filamentous bacteria have not been examined previously. In this case we found a typical actinomycete, Streptomyces sp. ES2-5, has ability to reduce Se(IV) to Se(0) and forms SeNPs in cells. TEM and EDX analyses showed that red-colored SeNPs accumulated in the hyphae with a diameter range of 50–500 nm. These bigger particles were aggregated by small SeNPs and then arranged along with hyphal cytoplasm as particle chains (Figs. 2, 3). It does not seem possible that these large Se(0) particles in the cytoplasm could have been derived from primary cytoplasmic synthesis and met cellular assimilation. Such a system for reduction of Se(VI) to Se(0) would be a detoxification mechanism. This mechanism could result in an incomplete selenite reduction under oxic growth conditions during a limited time frame (Fig. 4), which is consistent with a previous study in C. testosteroni [29]. Moreover, the large Se(0) particle chains could be extremely unmatched for hyphae, and thus the particle chains should be released only upon hyphal lysis or fragmentation (Fig. 2d, f). This could be very easy for filamentous bacteria due to the hyphal extending and branched growth. Similarly, the release of large Se(0) particles from cytoplasm via cell lysis can be observed in single-celled bacteria such as Bacillus mycoides [30] and B. selenitireducens [10]. In comparison with single-celled bacteria, it seems that Streptomyces sp. ES2-5 was lack of the mechanism of SeNP size control. In most cases, the diameters of SeNPs were <300 nm [4]. The size of SeNPs was about 200 nm even in filamentous fungi [40]. In Streptomyces sp. ES2-5, the size of larger SeNPs reached 500 nm (Fig. 3). The large Se(0) particles may also been formed by the aggregation of small particles during the movement of the cytoplasmic flow, which cannot be processed in single-celled bacteria and in eukaryotic cells with functional zoning.
As the diverse reducing mechanisms of Se oxyanions, it is very different from the reduction of As (such as ars cluster) and S (dsr cluster) associated with a certain enzymatic system in various bacteria [8, 41]. There could be multiple Se-reducing pathways in a strain, e.g., at least two selenite reductases in P. seleniipraecipitans or in R. selenitireducens [28, 31]. In Shewanella oneidensis MR-1, only 60 % selenite was reduced by reductase FccA [26], suggesting that more pathways are responsible for Se(IV) reduction in a bacterial strain. Among these reducing determinants, reduced thiols could be involved in Se(IV) reduction at more or less extent. Three bacterial groups produce thiols encompassing glutathione (GSH) in proteobacteria, bacillithiol (BSH) in Firmicutes and mycothiol (MSH) in actinobacteria [42]. As a result, it is not surprising that the most Se reducing bacteria distributed in these thiols-rich groups. Although a few studies on Se(IV) reduction in actinobacteria [38, 39], more Se(IV)-reducing actinobacteria would be examined in the future.
In this study, no red-colored precipitation was observed in different sub-cellular fractions in presence of NADPH or NADH as electron donors, suggesting NADPH or NADH dependent Se(IV) reductase was not responsible for Se(IV) reduction compared with previous studies [29–31]. Moreover, the intracellular concentration of GSH (could be MSH, an analog of GSH) per biomass decreased with Se(IV) reduction (Fig. 5), i.e., the reduced intracellular GSH was consumed to reduce Se(IV). In contrast, there were more extracellular thiols in broth amended with selenite than in control (Fig. 5b), indicating extracellular GSH was not involved in Se(IV) reduction because of its oxidized state under aerobic condition. Accordingly, the actinobacteria specific MSH (analog of GSH) was possibly involved in Se reduction in Streptomyces sp. ES2-5. Although the red-colored Se(0) nanoparticles are confirmed by Se(IV) reduction, this fact does not exclude the possibility that there are additional reduced products such as selenides because Streptomyces sp. ES2-5 has the ability to produce H2S. Se(IV) or Se(0) also might be reduced to Se(-II) via metabolic pathway of H2S production.
Many aerobic Se-oxyanions reducing bacteria were isolated from Enshi where soil has a high content of selenium [43–45]. Diversely aerobic Se-oxyanions reducing bacteria were also collected from different terrestrial soils [4, 13, 14, 29, 30]. This is different from the aquatic environments where anaerobic bacteria are responsible for Se(VI)/Se(IV) reduction. In anaerobic bacteria, Se(VI)/Se(IV) reduction is able to process on the surface of cells which is similar to Fe(III)’s reduction in S. oneidensis MR-1 [26]. In contrast, it is a great challenge in aerobic bacteria to reduce Se-oxyanions on surface of cells due to oxygen prior to accept the electrons than Se(IV), or other reducing determinants under oxidized stress in extracellular environment. Consequently, reduction is mainly processed in cells and then Se(0)/SeNP is exported or released by cell lysis (in this case).
Recent studies showed SeNPs synthesized by Streptomyces spp. had the anticancer activity [38, 39]. It implies the potential application of SeNPs from actinobacteria. In addition, bioremediation of contaminated soils needs aerobic microbes and anaerobic microbes. Streptomyces sp. ES2-5 not only has ability to reduce soluble Se(IV) into insoluble and less toxic SeNPs and to produce branched hyphae and countless spores, but also can adapt to selenite or chromate contaminated condition. Its ability of reproduction and adaptation in soil would be useful in Se/Cr bioremediation if Streptomyces sp. ES2-5 was applied to the contaminated site together with other aerobic and anaerobic Se(IV)-reducing bacteria.
Conclusions
A filamentous bacterium Streptomyces sp. was involved in reduction of Se(IV) to elemental SeNPs arranged in linear chains in cells under aerobic condition. The synthetic process of SeNPs and mechanism of Se(IV) reduction were proposed. The sizes of intracellular SeNPs varied from 50 to 500 nm and small SeNPs may aggregate into bigger particles. The cellular concentrations of GSH per biomass decreased along with Se(IV) reduction and Se(IV)-reduction did not occur in different sub-cellular fractions, showing that Se(IV) reduction was most possibly mediated by GSH in the cytoplasm, and thus the SeNPs were released via cell lysis or fragmentation.
Methods
The isolation, morphological and partial biochemical characters of strain ES2-5
The strain ES2-5 was isolated from a selenium mine soil (30°17′54″ N, 109°28′16″ E) with 38 mg kg−1 of total Se content in Hubei province, China and by serial dilutions of the sample on 1/10 TSA (tryptic soy agar, pH 7.3, Difco) containing 1 mM sodium selenite. After 2 days of incubation at 28 °C, colonies that developed a reddish color on the initial isolation plates were transferred to fresh media for further isolation, research and storage.
Strain ES2-5 was inoculated in 1/10 TSA plates supplemented with 10 mM sodium selenite. The plates without sodium selenite were served as controls. Then, the medium in plates was inserted with sterile glass slides at a diagonal angle and placed at 28 °C. After 3 days of cultivation, the cultures on the glass slide were fixed and stained using crystal violet for 2 min and then washed by water. After air-dried, the samples were observed using light microscopy.
Lecithinase enzyme activity, motility, hemolytic reaction, anaerobic growth, utilization of citrate, hydrolysis of gelation and tyrosine were tested using the conventional method. Production of H2S was tested according to the method described in [46].
16S rRNA gene sequencing and phylogenetic tree construction
The nearly-full 16S rRNA gene sequence of strain ES2-5 was amplified using 16S rDNA universal primers 27F and 1492R following the genomic DNA extraction. The accurate sequence of PCR product was acquired by sequencing after T-A cloning with a pGEM-T Easy vector (Promega). The 16S rRNA gene sequence was compared with sequences available in the EzTaxon-e server [47], and aligned with its close relatives using the CLUSTAL_X program [48]. Neighbor-joining tree was reconstructed using MEGA version 5.0 software [49]. Distances were calculated based on Kimura’s two-parameter method [50] and bootstrap analysis was performed according to 1000 resamplings [51]. The 16S rRNA gene sequence was registered as accession KF885787 in the GenBank database.
Growth, selenite resistance and reduction, and glutathione (GSH) determination
In order to determine the minimal inhibitory concentrations (MICs), strain ES2-5 was inoculated in 1/10 TSA plates with different concentrations of Se(IV) (0, 1, 5, 10, 20, 50, 100, 150 mM) and Cr(VI), Zn(II), Cu(II), As(III) and Sb(III) at 28 °C.
The growth curve was determined by inoculating strain ES2-5 into 100 ml 1/10 TSB broth supplemented without or with 1.0 mM sodium selenite at 28 °C with shaking at 160 rpm. Cultures were taken at 4 h intervals and centrifuged at 6000×g, 5 min. The supernatants (a) were used to determine concentrations of extracellular GSH and selenite. The pellet was washed twice with phosphate buffer saline (PBS, pH 7.2) and re-suspended in the same buffer. Then, the suspension was sonicated for 3 min and centrifuged at 6000×g and 4 °C for 5 min. The supernatants (b) were collected and used to measure the concentrations of intracellular GSH and totally cellular proteins. The determination for the content of GSH was performed by using a fluorescence-based method as described in [52]. The detailed process was realized as follows. 10 mM naphthalene-2,3-dicarboxaldehyde (NDA) was dissolved in dimethylsulfoxide (DMSO) and 50 mM Tris–HCl (pH 10.0). The NDA reagent reacts with amino and sulfhydryl groups of GSH to form an adduct, which can be measured by fluorescence signal (λ exc at 472 nM and λ em at 528 nM). The standard curve of the relationship between concentrations of GSH and the value of fluorescence signal was measured by using 100, 200, 300, 400 and 500 nM reduced GSH standard solution. The GSH concentrations of samples were calculated according to standard curve and detected fluorescence value. The biomass of strain ES2-5 was tested by measuring the contents of total cellular proteins of samples using Coomassie brilliant blue G-250 method with Bovine Serum Albumin (BSA) as standard [53]. Selenite concentrations in the supernatants (a) were measured by HPLC-HG-AFS (Beijing Tian Instruments Co., Ltd., China) [54].
Selenite reduction activity assays in cultural supernatant and cellular fractions
In order to determine the ability of vitro Se(IV) reduction by cultural supernatant and different cellular fractions, the culture was grown to log phase and centrifuged at 6000×g, 5 min. The cultural supernatant was collected and filtered by a filtration with 0.2 µm disks. The pellet was washed twice with phosphate buffer saline (PBS, pH 7.2) and resuspended in the same buffer for sonication. After sonication for 3 min, the cell lysate was centrifuged at 6000×g for 5 min to remove the cell debris. Then, the soluble supernatant was centrifuged at 20,000×g for 60 min to separate the cytoplasmic fraction and membrane fraction. Selenite reductase activity was determined using the following reaction mixture [29, 30]: cultural supernatant, cytoplasmic or membrane fraction; sodium selenite (final concentration 0.2 mM); NADPH or NADH (final concentration 0.2 mM). The reaction mixture was incubated at 28 °C for 24 h. Reaction mixture without addition of cultural supernatant, cytoplasmic or membrane fraction served as controls. Selenite concentrations in the reaction mixture were measured by HPLC-HG-AFS (Beijing Titan Instruments Co., Ltd., China) [54].
Scanning electron microscopy (SEM)
Strain ES2-5 was grown in TSB supplemented without or with 1.0 mM sodium selenite at 28 °C, 160 rpm. After 3 days of cultivation, cells were centrifuged (6000 rpm, 10 min, 4 °C) and scanning electron microscopic observation was performed on the processed samples. Samples processing involves washing, fixing and drying of cells. Harvested cells were washed thrice with phosphate buffer saline (PBS, pH 8.0). Fixation was conducted with 2.5 % glutaraldehyde (24 h, 4 °C). Cells were washed again with PBS. Fixed cells were dehydrated through a series of alcohol dehydration steps (30, 50, 70, 80, 90 and 100 %) and finally freeze dried and sputter coated. The samples were then viewed using SEM (JSM-6390 JEOL JAPAN). Samples collected from the culture without addition of selenite were regarded as controls.
Transmission electron microscopy (TEM) and SeNP analysis with energy dispersive X-ray (EDX)
To obtain ultra-thin sections for TEM and EDX analysis, harvested cells through above-mentioned method were fixed using 2 % v/v glutaraldehyde in 0.05 M sodium phosphate buffer (pH 7.2) for 24 h and were then rinsed three times in 0.15 M sodium cacodylated buffer (pH 7.2) for 2 h. The specimens were dehydrated in graded series of ethanol (70, 96 and 100 %) transferred to propylene oxide and embedded in Epon according to standard procedures. The sections, approximately 80 nm thick, were cut with an ultrathin E (Reichert Jung) microtome and collected on copper grids with Formvar supporting membranes. The sections were stained with uranyl acetate and lead citrate and then TEM-EDX (JEM2100F JAPAN) was performed.
Abbreviations
- EDX:
-
electron dispersion spectroscopy
- MIC:
-
minimal inhibitory concentration
- SEM:
-
scanning electron microscopy
- SeNPs:
-
selenium nanoparticles
- TEM:
-
transmission electron microscopy
References
Hatfield DL, Berry MJ, Gladyshev VN. Selenium: Its molecular biology and role in human health. 3rd ed. London: Springer science+business media; 2012.
Soboh B, Pinske C, Kuhns M, Waclawek M, Ihling C, Trchounian K, Trchounian A, Sinz A, Sawers G. The respiratory molybdoselenoprotein formate dehydrogenases of Escherichia coli have hydrogen:benzyl viologen oxidoreductase activity. BMC Microbiol. 2011;11:173.
Shaw FL, Mulholland F, Gall GL, Porcelli I, Hart DJ, Pearson BM, Van Vliet AHM. Selenium-dependent biogenesis of formate dehydrogenase in Campylobacter jejuni is controlled by the fdhTU accessory genes. J Bacteriol. 2012;194:3814–23.
Nancharaiah YV, Lens PNL. Ecology and biotechnology of selenium-respiring bacteria. Microbiol Mol Biol Rev. 2015;79:61–80.
Winkel LH, Johnson CA, Lenz M, Grundl T, Leupin OX, Amini M, Charlet L. Environmental selenium research: from microscopic processes to global understanding. Environ Sci Technol. 2012;46:571–9.
Combs JF Jr. Selenium in global food systems. Br J Nutr. 2001;85:517–47.
Dowdle PR, Oremland RS. Microbial oxidation of elemental selenium in soils lurries and bacterial cultures. Environ Sci Technol. 1998;32:3749–55.
Stolz JF, Basu P, Santini JM, Oremland RS. Arsenic and selenium in microbial metabolism. Annu Rev Microbiol. 2006;60:107–30.
Kessi J, Ramuz M, Wehrli E, Spycher M, Bachofen R. Reduction of selenite and detoxification of elemental selenium by the phototrophic bacterium Rhodospirillum rubrum. Appl Environ Microbiol. 1999;65:4734–40.
Oremland RS, Herbel MJ, Blum JS, Langley S, Beveridge TJ, Ajayan PM, Sutto T, Ellis AV, Curran S. Structural and spectral features of selenium nanospheres produced by Se-respiring bacteria. Appl Environ Microbiol. 2004;70:52–60.
Kessi J. Enzymic systems proposed to be involved in the dissimilatory reduction of selenite in the purple non-sulfur bacteria Rhodospirillum rubrum and Rhodobacter capsulatus. Microbiology. 2006;152:731–43.
Hunter WJ, Kuykendall LD. Reduction of selenite to elemental red selenium by Rhizobium sp. strain B1. Curr Microbiol. 2007;55:344–9.
Bajaj M, Schmidt S, Winter J. Formation of Se (0) Nanoparticles by Duganella sp. and Agrobacterium sp. isolated from Se-laden soil of North-East Punjab, India. Microb Cell Fact. 2012;11:115–20.
Lampis S, Zonaro E, Bertolini C, Cecconi D, Monti F, Micaroni M, J.Turner R, S.Butler C, Vallini G. Selenite biotransformation and detoxification by Stenotrophomonas maltophilia SeITE02: novel clues on the route to bacterial biogenesis of selenium nanoparticles. J Haz Mat. 2016. http://dx.doi.org/10.1016/j.jhazmat.2016.02.035.
Forootanfara H, Mahboubeh A, Maryam N, Mitra M, Bagher A, Ahmad S. Antioxidant and cytotoxic effect of biologically synthesized selenium nanoparticles in comparison to selenium dioxide. J Trace Elem Med Biol. 2014;28(1):75–9.
Hariharan H, Al-harbi N, Karuppiah P, Rajaram S. Microbial synthesis of selenium nanocomposite using Saccharomyces cerevisiae and its antimicrobial activity against pathogens causing nosocomial infection. Chalcogenide Lett. 2012;9:509–15.
Yang F, Tang Q, Zhong X, Bai Y, Chen T, Zhang Y, Li Y, Zhang X. Surface decoration by Spirulina polysaccharide enhances the cellular uptake and anticancer efficacy of selenium nanoparticles. Int J Nanomedicine. 2012;7:835–44.
Yazdi MZ, Mahdavi M, Varastehmoradi B, Faramarzi MA, Shahverdi AR. The immunostimulatory effect of biogenic selenium nanoparticles on the 4T1 breast cancer model: an in vivo study. Biol Trace Elem Res. 2012;149:22–8.
Wang T, Yang L, Zhang B, Liu J. Extracellular biosynthesis and transformation of selenium nanoparticles and application in H2O2 biosensor. Colloids Surf. B. 2010;80:94–102.
Zhang J, Zhang S, Xu J, Chen H. A new method for the synthesis of selenium nanoparticles and the application to construction of H2O2 biosensor. Chin Chem Lett. 2004;15:1345–8.
Losi M, Frankenberger W. Reduction of selenium oxyanions by Enterobacter cloacae SLD1a-1: isolation and growth of the bacterium and its expulsion of selenium particles. Appl Environ Microbiol. 1997;63:3079–84.
Jiang S, Cuong T, Lee J, Duong H, Han S, Hur H. Mercury capture into biogenic amorphous selenium nanospheres produced by mercury resistant Shewanella putrefaciens. Chemosphere. 2012;87:621–4.
Fellowes J, Pattrick R, Green D, Dent A, Lloyd J, Pearce C. Use of biogenic and abiotic elemental selenium nanospheres to sequester elemental mercury released from mercury contaminated museum specimens. J Hazard Mater. 2011;189:660–9.
DeMoll-Decker H, Macy JM. The periplasmic nitrite reductase of Thauera selenatis may catalyze the reduction of selenite to elemental selenium. Arch Microbiol. 1993;160:241–7.
Hunter WJ, Kuykendall LD. Identification and characterization of an Aeromonas salmonicida (syn Haemophilus piscium) strain that reduces selenite to elemental red selenium. Curr Microbiol. 2006;52:305–9.
Li DB, Cheng YY, Wu C, Li WW, Li N, Yang ZC, Tong ZH, Yu HQ. Selenite reduction by Shewanella oneidensis MR-1 is mediated by fumarate reductase in periplasm. Sci Rep. 2014;4:3755.
Antonioli P, Lampis S, Chesini I, Vallini G, Rinalducci S, Zolla L, Righetti PG. Stenotrophomonas maltophilia SeITE02, a new bacterial strain suitable for bioremediation of selenite-contaminated environmental matrices. Appl Environ Microbiol. 2007;73:6854–63.
Hunter WJ. Pseudomonas seleniipraecipitans proteins potentially involved in selenite reduction. Curr Microbiol. 2014;69:69–74.
Zheng S, Su J, Wang L, Yao R, Wang D, Deng Y, Wang R, Wang G, Rensing C. Selenite reduction by the obligate aerobic bacterium Comamonas testosteroni S44 isolated from a metal-contaminated soil. BMC Microbiol. 2014;14:204–16.
Lampis S, Zonaro E, Bertolini C, Bernardi P, Butler CS, Vallini G. Delayed formation of zero-valent selenium nanoparticles by Bacillus mycoides SelTE01 as a consequence of selenite reduction under aerobic conditions. Microb Cell Fact. 2014;13:106–11.
Hunter WJ. A Rhizobium selenitireducens protein showing selenite reductase activity. Curr Microbiol. 2014;68:311–6.
Hockin SL, Gadd GM. Linked redox precipitation of sulfur and selenium under anaerobic conditions by sulfate-reducing bacterial biofilms. Appl Environ Microbiol. 2003;69:7063–72.
Painter EP. The chemistry and toxicity of selenium compounds with special reference to the selenium problem. Chem Rev. 1941;28:179–213.
Kessi J, Hanselmann KM. Similarities between the abiotic reduction of selenite with glutathione and the dissimilatory reaction mediated by Rhodospirillum rubrum and Escherichia coli. J Biol Chem. 2004;279:50662–9.
Afkar E, Lisak J, Saltikov C, Basu P, Oremland RS, Stolz JF. The respiratory arsenate reductase from Bacillus selenitireducens strain MLS10. FEMS Microbiol Lett. 2003;226:107–12.
Yanke LJ, Bryant RD, Laishley EJ. Hydrogenase (I) of Clostridium pasteurianum functions a novel selenite reductase. Anaerobe. 1995;1:61–7.
Tomei FA, Barton LL, Lemanski CL, Zocco TG, Fink NH, Sillerud LO. Transformation of selenate and selenite to elemental selenium by Desulfovibrio desulfuricans. J Ind Microbiol. 1995;14:329–36.
Ahmad MS, Yasser MM, Sholkamy EN, Ali AM, Mehanni MM. Anticancer activity of biostabilized selenium nanorods synthesized by streptomyces bikiniensis strain Ess_amA- 1. Int J Nanomed. 2015;10:3389–401.
Ramya S, Shanmugasundaram T, Balagurunathan R. Biomedical potential of actinobacterially synthesized selenium nanoparticles with special reference to anti-biofilm, anti-oxidant, wound healing, cytotoxic and anti-viral activities. J Trace Elem Med Bio. 2015;32:30–9.
Vetchinkina E, Loshchinina E, Kursky V, Nikitina V. Reduction of organic and inorganic selenium compounds by the edible medicinal Basidiomycete Lentinula edodes and the accumulation of elemental selenium nanoparticles in its mycelium. J Microbiol. 2013;51:829–35.
Venceslau SS, Stockdreher Y, Dahl C, Pereira IAC. The “bacterial heterodisulfide” dsrc is a key protein in dissimilatory sulfur metabolism. BBA-Bioenergetics. 2014;1837:1148–64.
Fahey RC. Glutathione analogs in prokaryotes. BBA-Bioenergetics. 1830;2013:3182–98.
Yao R, Wang R, Wang D, Su J, Zheng S, Wang G. Paenibacillus selenitireducens sp. nov., a selenite-reducing bacterium isolated from a selenium mineral soil. Int J Syst Evol Microbiol. 2014;64:805–11.
Xiang W, Wang G, Wang Y, Yao R, Zhang F, Wang R, Wang D, Zheng S. Paenibacillus selenii sp. nov., isolated from selenium mineral soil. Int J Syst Evol Microbiol. 2014;64:2662–7.
Li X, Kot W, Wang D, Zheng S, Wang G, Hansen LH, Rensing C. Draft genome sequence of Se(IV)-reducing bacterium Pseudomonas migulae ES3-33. Gen Announc. 2015;3(3):e00406–15.
Dong XZ, Cai MY. Determinative manual for routine bacteriology. Beijing: Scientific Press; 2001.
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH. Introducing EzTaxon-e:a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–21.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9.
Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20.
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91.
Lewicki K, Marchand S, Matoub L, Lulek J, Coulon J, Leroy P. Development of a fluorescence-based microtiter plate method for the measurement of glutathione in yeast. Talanta. 2006;70(4):876–82.
Van Wilgenburg MG, Werkman EM, Van Gorkom WH, Soons JB. Criticism of the use of Coomassie Brilliant Blue G-250 for the quantitative determination of proteins. J Clin Chem Clin Biochem. 1981;19:301–4.
Li J, Wang Q, Zhang SZ, Qin D, Wang GJ. Phylogenetic and genome analyses of antimony-oxidizing bacteria isolated from antimony mined soil. Int Biodeter Biodegr. 2013;76:76–80.
Authors’ contributions
SZ and GW designed the experiments. YT and RY conducted the experiments. DW and RW assisted to the GSH determination. SZ and YT analyzed the results and wrote the manuscript. GW reviewed and revised the manuscript. All authors have read and approved the final manuscript.
Acknowledgements
We thank Dr. Qin at the Electronic Microscope Center of Huazhong Agricultural University.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Funding
This work was supported by the National Natural Science Foundation of China (31470227) and the fund of the Tobacco Company of Enshi, Hubei Province, P. R. China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Yuanqing Tan and Rong Yao contributed equally to this work
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Tan, Y., Yao, R., Wang, R. et al. Reduction of selenite to Se(0) nanoparticles by filamentous bacterium Streptomyces sp. ES2-5 isolated from a selenium mining soil. Microb Cell Fact 15, 157 (2016). https://doi.org/10.1186/s12934-016-0554-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12934-016-0554-z