Abstract
Background
Babesia spp. are tick-borne protozoan hemoparasites and the second most common blood-borne parasites of mammals, in particular domestic animals. We used the Clustal Multiple Alignment program and 18S rRNA gene sequences of 22 Babesia species from GenBank to develop a PCR that could detect a wide variety of Babesia spp. in a single reaction. The pan-Babesia FRET-qPCR we developed reliably detected B. gibsoni, B. canis, B. vogeli, B. microti, B. bovis, and B. divergens under controlled conditions but did not react with closely related species, mainly Hepatozoon americanum, Theileria equi, and Toxoplasma gondii.
Results
When we tested the pan-Babesia FRET-qPCR on DNA of whole blood from 752 cattle, sheep, goats, donkeys and horses from five Caribbean islands, we detected Babesia spp. expected to be present in the animals, mainly B. bovis and B. bigemina in cattle and B. caballi in horses and donkeys. Further, we found that animals were not uncommonly infected with species of Babesia usually associated with other hosts, mainly B. vogeli and B. gibsoni in cattle, sheep and goats, B. rossi in goats, and B. caballi in goats and sheep. Finally, the pan-Babesia FRET-qPCR enabled us to identify unknown species of Babesia in cattle, goats, sheep and donkeys.
Conclusions
Overall, 70 % (525/752) of the animals we tested were positive confirming earlier limited studies that infections with Babesia spp. are common in livestock in the Caribbean.
Similar content being viewed by others
Background
Babesia spp. are tick-borne protozoan hemoparasites that occur worldwide [1–4]. They are the second most common blood-borne parasites of mammals, after trypanosomes, with infections occurring commonly in domestic animals, in particular cattle, dogs, horses, sheep, and pigs [5]. Recently, infections with Babesia (babesiosis) have been described in birds [6–8] and have attracted increasing attention as zoonotic infections in people [5, 9].
Since the first description of Babesia in cattle by Victor Babes in 1888, over 100 Babesia species have been identified [8]. Many cause significant economic losses in livestock, mainly B. bovis and B. bigemina in cattle [10], B. motasi and B. ovis in small ruminants [11] and B. caballi in horses and donkeys [12]. Further, B. canis, B. vogeli and B. gibsoni are important causes of morbidity and mortality in dogs worldwide [13] while B. microti and B. divergens are the species that most commonly infect people [8].
Initially, differentiation of the Babesia spp. was based on morphological and biological characteristics, and invertebrate and vertebrate host specificity. With the advent of molecular tools, however, these methods have proven to be of limited taxonomic value [8]. A number of nucleic acid-based techniques have been reported which detect Babesia spp. with high sensitivity and specificity. Most commonly, these assays have a narrow spectrum and specifically identify B. microti, B. divergens, or groups of Babesia spp. associated with specific hosts such as dogs [14, 15], cattle [16, 17] or sheep [18, 19]. To enable the detection of a wide range of Babesia spp. of veterinary and public health significance in a single reaction we developed a broad-based qPCR. Further, we tested our pan-Babesia FRET-qPCR on DNAs extracted from whole blood samples collected from five livestock species on five Caribbean islands. The results of these experiments are described below.
Methods
Whole blood
Whole blood samples (n = 752) were collected into EDTA from apparently healthy livestock on five Caribbean islands, including 162 from Dominica (cattle = 77, goats = 70, and sheep = 15), 31 from Grenada (all goats), 93 from Montserrat (cattle = 12, goats = 19, and sheep = 62), 198 from Nevis (cattle = 43, goats = 114, and sheep = 41) and 268 from St. Kitts (cattle = 193, goats = 4, sheep = 26, donkeys = 25, and horses = 20) [20]. The study was reviewed and approved by the Institutional Animal Care and Use Committee of the Ross University School of Veterinary Medicine (RUSVM), St Kitts.
DNA extraction
After collection, the blood samples were transported on ice to RUSVM where red blood cells were separated by centrifugation and stored at −20 °C until thawed at room temperature and DNA extracted from aliquots (200 μL) using the QIAamp DNA Blood Mini Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions. The DNA was eluted in 200 μL washing buffer and shipped to Yangzhou University College of Veterinary Medicine of Jiangsu province, China at room temperature where it was frozen at −80 °C until PCRs were performed.
Pan-Babesia FRET-qPCR
The PCRs in this study were performed on a Roche Light-Cycler 480-II platform. The HMBS-based quantitative PCR was used as an endogenous quality control to verify the quality of the DNA in the samples [21].
Primers and probes
The 18S rRNA sequences for 22 recognized Babesia spp. of public health significance and/or veterinary importance were obtained from GenBank (Fig. 1): B. microti (AB071177, AB219802), B. leo (AF244911, AY452708), B. rodhaini (DQ641423, AB049999), B. felis (AF244912, AY452707), B. poelea (DQ200887), B. bigemina (JQ723014, KF112076), B. bovis (HQ264124, HQ264127), B. caballi (AY534883), B. canis (AY072926, JN982353), B. capreoli (FJ944828, GQ304526), B. crassa (AY260176, JX542614), B. divergens (FJ944822, FJ944826), B. gibsoni (KJ142323), B. hongkongensis (JQ867356), B. kiwiensis (EF55315), B. major (JF802040), B. motasi (AY260179, AY533147), B. odocoilei (AY661508, U16369), B. ovata (AY081192, AY603400), B. rossi (JN982353), B. vitalii (JN880430, JN880431) and B. vogeli (HM590440). In addition, the 18S rRNA sequences of 9 related protozoan species were also obtained from GenBank: Theileria equi (AB515307, AB515312), T. parva (L02366), Trypanosoma cruzi (AF303659), Toxoplasma gondii (L37415), Neosporo caninum (U63069), Eimeria arnyi (AY613853), Cytauxzoon felis (AY679105), Hepatozoon americanum (AF176836), Cryptosporidium meleagridis (AF112574) and C. parvum (L16996).
Alignment of the partial 18S rRNA gene amplicons of Babesia spp. and other related species. The upstream primer-1 (in red), upstream primer-2 (in blue), the fluorescein/LCRed 640 probes and the downstream primer are indicated in the top of the boxes. Dots indicate nucleotides identical to the primers and probes, and the dashes denote the deletion of the nucleotides. The upstream primers and two probes are used as the indicated sequences while the downstream primer is used as antisense oligonucleotide. While the probes and downstream primer show minimum mismatch with Babesia spp. and other related species, the upstream primers (−1 and −2) have 0–1 nucleotide mismatch with Babesia spp. but 6–16 nucleotide mismatches with the related non- Babesia species
These sequences were aligned using Clustal Multiple Alignment to identify conserved and variable regions suitable for primers and probes that could differentiate the species. The upstream primer-1 (5′-ATG GCT TTG CCG GCG ATG TAT CA-3′) was selected because of its high specificity for five Babesia species while a second upstream-2 primer (5′- TTT HGC GAT GKW CCA TTC AAG TTT CTG -3′) was selected because it reacted with 17 other Babesia species. Both had multiple mismatches with other related protozoan species (Fig. 1). The reverse primer (5′- CTG GCA CCA GAC TTG CCC TCC AAT -3′), the fluorescein probe [5′- ACG GGT AAC GGG GAA TTA GGG TTC-(6-FAM)-3′] and LCRed 640 probe [5′-(LCRed 640)-ATT CCG GAG AGG GAG CCT GAG AAA-PHOS-3′] were selected as they had only few mismatches amongst the Babesia but high numbers of mismatches with the related species we studied (Fig. 1).
Thermal cycling and melting curve analysis
The pan-Babesia FRET-qPCR was performed in a LighCycler 480-II real-time PCR platform using conditions described preciously [22] with a hybridization temperature of 58 °C. The PCR master mix contained the two upstream primers, the downstream primer, and two probes in concentrations described previously [23]. High-resolution melting curve analysis was performed following the completion of PCR [22]. Data were analyzed as 640 nm: 530 nm (F4/F1) fluorescence ratios, and the first derivative of F4/F1 (−d(F4/F1)/dt) was evaluated.
Sensitivity and specificity
To test the specificity of the pan-Babesia FRET-PCR, we used DNAs obtained in previous studies [20, 24] of B. gibsoni, B. canis, B. vogeli as positive controls and DNAs from Hepatozoon americanum, Theileria equi and Toxoplasma gondii as negative controls. In addition, we used plasmids created to contain the 18S rRNA gene (Integrated DNA Technologies, Coralville, IA, USA) of B. microti, B. bovis, and B. divergens as positive controls and T. equi as negative controls.
To test the sensitivity of the pan-Babesia FRET-PCR we used quantitative standards consisting of amplification products of PCRs for B. gibsoni, B. canis and B. vogeli identified in a previous study [14, 22]. The amplicons were gel purified with the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA), quantified by the PicoGreen DNA fluorescence assay (Molecular Probes, Eugene, OR), and sequenced at the Genomic Sequencing Laboratory (GenScript, Nanjing, Jiangsu, China). The purified amplicons were diluted at 1,000, 100, 10, 1 genome copies per PCR reaction in T10E0.1 buffer as described previously [24], and used as quantitative standards.
In the specificity and sensitivity tests, the PCR products were electrophoresed through 1.5 % MetaPhor agarose gels. We calculated the size of the possible PCR amplicons for the different Babesia species to be between 282 to 293 bp and random samples with products within this range were purified for automated DNA sequencing (GenScript, Jiangsu, Nanjing, China) with a QIAquick PCR Purification Kit according to the manufacturer’s instructions (Qiagen, Valencia, CA, USA). The sequencing was performed by GenScript (Jiangsu, Nanjing, China) using the upstream and downstream primers to determine the Babesia species. In cases where there were poorly defined or multiple peaks in the sequencing results indicating mixed infections, pGEM®-T Easy Vector Systems (Promega, Madison, WI, USA) was used to clone PCR amplicons following the manufacturer’s instructions.
The 18S rRNA gene has been widely used to determine phylogenetic relationships between Babesia spp. [8] and it is generally accepted that the rRNA gene sequence similarity between two strains of the same species is over 98.65 % [25–27]. If the rRNA sequence for the Babesia isolate we obtained had a similarity of over 98.65 % with a Babesia species on GenBank, the isolate was regarded as being that species (Table 2).
Confirmatory nested PCR
Nested primers (Outer primers: upstream: 5′-CATCAGCTTGACGGTAGGGTATT-3′, downstream 5′-CCCCCAACCGTTCCTATTAAC-3′; amplicon size: 489–518 bp; Inner primers: upstream: 5′-GAGGCAGCAACGGGTAACG-3′, downstream 5′-CCAACAAAATAGAACCAAAGTCCTA-3′; amplicon size: 421–447 bp) were designed to target a hyper-variable region of the 18S rRNA gene to amplify samples found positive for Babesia by FRET-PCR.
Results
Establishment of the pan-Babesia FRET-qPCR
The primers and probes we designed had 0–3 nucleotide mismatches with the 22 Babesia spp. with which they were compared, but 6–19 mismatches with 9 other related protozoan species (Fig. 1). The generic pan-Babesia FRET-qPCR we developed specifically detected DNAs of B. gibsoni, B. canis, B. vogeli, and plasmids from B. microti, B. bovis, B. divergens, but not DNAs from H. americanum, T. equi and T. gondii, and plasmids from T. equi. Using the purified Babesia DNAs (B. canis, B. gibsoni and B. vogeli) as quantitative standards, we determined that the detection limit of the FRET-qPCR was ~2 copies of 18S rRNA gene per PCR reaction, equivalent to 20 copies of 18S rRNA per ml whole blood. The Babesia-positive samples based on FRET-qPCR were verified in confirmatory nested PCRs and by sequence determination of the amplification products for precise species identification.
Prevalence of Babesia spp. in Caribbean islands
Of the 752 blood samples we examined, 525 (70 %) were positive for Babesia spp. in our pan-Babesia FRET-qPCR. Although numbers were small, donkeys (88 %; 22/25) and horses (80 %; 16/20) were most commonly positive for Babesia spp. followed by cattle (78 %; 274/352), sheep (70 %; 101/144), and goats (47 %; 112/238). The results indicate Babesia spp. occur widely in the region with evidence of infection found on all islands studied, mainly Nevis (46 %; 92/198), Dominica (56 %; 90/162), Grenada (74 %; 23/31), and Montserrat (94 %; 87/93) St. Kitts (87 %; 233/268).
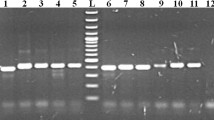
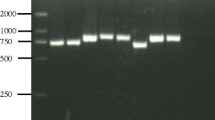
The PCR amplicons (282–293 bp) of our pan-Babesia FRET-qPCR were polymorphic among the different Babesia species and genomic sequencing thus enabled us to determine the species of Babesia amplified. Of the 525 Babesia-positive samples detected by the pan-Babesia FRET-qPCR, amplicons from 84 (84/525, 16 %) were randomly selected and sequenced revealing seven Babesia species were present in the livestock we studied from five Caribbean islands (Table 1, Fig. 2). These were B. bigemina (17/84, 20 %), B. bovis (6/83, 7 %), B. caballi (20/83, 24 %), B. vogeli (21/83, 25 %), B. gibsoni (8/83, 10 %), B. rossi (1/83, 1 %) and 11 unnamed Babesia spp. (11/83, 13 %).
Phylogenetic comparison of the 282–293 bp variable region of the 18S rRNA gene found in the Babesia we found in our study (in red) and published Babesia sequences in GenBank (in black). Branch lengths are measured in nucleotide substitutions and numbers show branching percentages in bootstrap replicates
Cloning of PCR amplicons enabled us to identify dual infections with two or three Babesia species in 3 bovines from Montserrat (B. vogeli and B. bigemina), 1 bovines from Dominica (B. vogeli and B. bigemina), 1 goat from Montserrat (B. vogeli and Babesia sp.), 4 bovines from Dominica (B. vogeli and B. gibsoni), 2 bovines (B. vogeli and B. bigemina) and 1 sheep (B. vogeli and B. gibsoni) from St. Kitts, and 2 sheep from Montserrat (B. vogeli, B. caballi and an unnamed Babesia spp.).
While B. caballi was the main species identified by sequencing of positive amplicons from horses (5/5, 100 %) and donkeys (5/7, 71 %), up to four Babesia spp. were found in cattle, goats and sheep from the islands (Table 1). While DNA of B. bigemina and B. bovis were only found in cattle with the former being most prevalent (17/41, 41 %), DNA of B. vogeli and B. gibsoni was found in all the domestic ruminants. The DNA of B. vogeli was most commonly found in cattle (15/41, 37 %) while that of B. gibsoni was found most commonly found in sheep (5/18, 28 %). The DNA of B. caballi was not found in cattle but was found in sheep (4/18, 22 %) and also in goats (6/13, 46 %), where it was the most common species present. The DNA of B. rossi were found in only one goat while that of unnamed Babesia were found in all species on all islands but were most prevalent in sheep (6/18, 33 %; Table 1).
Discussion
The pan-Babesia FRET-qPCR we developed proved to be both specific and sensitive in detecting Babesia spp. in controlled experiments. In all positive control reactions it identified Babesia spp. correctly and failed to give amplicons with the negative control organisms that were closely related to Babesia (Fig. 1). The test could detect as few as 20 copies of the 18S rRNA gene per ml of whole blood indicating it would be useful in detecting chronic infections with low parasitemias that are common in infections with Babesia species. To the best of our knowledge, the pan-Babesia FRET-qPCR we designed is the first real-time PCR to be reported which detects multiple Babesia spp. with high sensitivity and specificity. We selected the 18S rRNA gene as the target for our FRET-qPCR as this is the most often used and reliable target gene for PCR detection of Babesia spp. [14]. Although the nucleotide sequences of the 18S rRNA gene are very similar amongst Babesia spp. and other related protozoan species, by systematically aligning the sequences of 32 Babesia and related species we were able to identify a highly conserved region specific for Babesia spp.. We developed specific primers (upstream primer-1 and upstream primer-2) to amplify this region and ensure that only Babesia spp. were detected and no related species. Further, the region of the 18S RNA gene we selected for our FRET-qPCR had nucleotide mismatches between the Babesia spp. we studied which enabled us to differentiate the organisms by sequencing the amplicons we obtained. When we compared the GenBank sequences of the 282–293 bp sections of the 18S rRNA gene of the recognized Babesia spp. detected by our pan-Babesia FRET-qPCR, we found each recognized Babesia sp. had 98.3 % or less similarity with the others (Table 2). This supports previous suggestions [25–27], based on comparisons of entire 18S rRNA gene sequences, that strains of organisms within the same species have similarities of over 98.65 %. When we applied our pan-Babesia FRET-qPCR to whole blood collected from five types of livestock from five Caribbean islands, we identified high prevalences of infections with Babesia species. This is not unexpected as the tick vectors of Babesia are common on livestock in the Caribbean [28, 29] and there are reports of high infection rates with B. bigemina, B. bovis and B. caballi [30–35]. The most prevalent Babesia species we found on cattle, B. bovis and B. bigemina, are transmitted by Rhipicephalus microplus which is very common in the Caribbean [29]. Although R. microplus has been found on sheep and goats in the Caribbean [28] and elsewhere [36] we found no evidence of transmission of B. bigemina or B. bovis in these small ruminants. We also found no evidence of the common small ruminant Babesia spp. mainly B. ovis, B. motasi and B. sp. Xinjiang [8] in the small ruminants we studied but this was not unexpected as their vectors, Rhipicephalus bursa, Hemaphysalis spp. and H. longicornis, respectively, do not occur in the region.
The commonest Babesia we found in the equids we studied was B. caballi, the agent of equine piroplasmosis which is common in the Caribbean [34]. There are at least 13 tick species incriminated in the transmission of B. caballi of which Dermacentor (Anocentor) nitens is considered to be the major vector in Latin America [37]. The tropical horse tick, D. nitens, is common on horses and donkeys in the Caribbean [28, 34] and can also be found on ruminants in the region [38] which might explain our findings of B. caballi in relatively high numbers of sheep and goats. Although we could find no reference to B. caballi infections in domestic ruminants, there is a report of a Babesia in desert bighorn sheep that had cross reacting antigens with B. caballi [39].
The traditional method of identifying Babesia species based on their size, numbers of daughter cells following merozoite division, and vertebrate and invertebrate host specificity have been replaced by molecular methods which have shown that Babesia have broader host ranges than thought previously [8]. This appeared to be the case in our study where we identified a number of Babesia not previously thought to infect livestock. The most common was B. vogeli which is a common cause of babesiosis in dogs around the world and also in the Caribbean [22, 40, 41]. Recently it has been reported in cats in Thailand [42] and lions in Zimbabwe [43], and our study extends its potential host range to domestic ruminants. The vector of B. vogeli is Rhipicephalus sanguineus which is found very commonly on dogs in the Caribbean [22, 40, 41]. While there are no reports of R. sanguineus on livestock in the Caribbean, the tick has been found occasionally on livestock elsewhere [44, 45]. Recently DNA of B. vogeli was found in Rhipicephalus turanicus in Israel [46] and this tick might be a vector of the organism. Rhipicephalus turanicus is morphologically very similar to R. sanguineus but is found on a wider host range including dogs, domestic ruminants and horses [47]. Further studies are indicated to determine if R. turanicus occurs in the Caribbean and if it is a vector of B. vogeli.
Babesia gibsoni is another agent of canine babesiosis which seems to be transmitted by the R. sanguineus group. The organism occurs widely, in northern Africa, southern Asia, Australia, Europe, the USA, the Caribbean and Central America [48], but ours is the first report of B. gibsoni in domestic ruminants. Similarly, although Babesia rossi was thought to be restricted to Africa where it is transmitted amongst dogs by Haemaphysalis elliptica [13], it has recently been reported in a Haemaphysalis parva in Turkey [49] and we now report the organism in a goat in the Caribbean. To the best of our knowledge, Haemaphysalis spp. have not been reported in the Caribbean and further studies are needed to determine the epidemiology of B. rossi in the area. As this organism is highly pathogenic in dogs it should be suspected in animals in the Caribbean that develop severe signs of canine babesiosis.
Conclusions
In conclusion, our experiments have shown that the pan-Babesia FRET-qPCR we developed can reliably detect a variety of Babesia spp. under controlled conditions. Further, when used on bloods from livestock on Caribbean islands, it enabled us to detect Babesia spp. that were expected to be present in the animals we studied and to support previous studies showing infections are common in livestock in the Caribbean. In contrast to morphological studies and parasite specific PCRs, the pan-Babesia FRET-qPCR enabled us to demonstrated that livestock are not uncommonly infected with Babesia spp. that are usually associated with other hosts and that host specificity of Babesia spp. is likely less than thought previously.
References
Bai Q, Liu G, Liu D, Ren J, Li X. Isolation and preliminary characterization of large Babesia sp. from sheep and goats in the eastern part of Gansu Province, China. Parasitol Res. 2002;13 Suppl 1:16–21.
Criado-Fornelio A, Martinez-Marcos A, Buling-Saraña A, Barba-Carretero JC. Molecular studies on Babesia, Theileria and Hepatozoon in southern Europe: Part II. Phylogenetic analysis and evolutionary history. Vet Parasitol. 2003;114:173–94.
Uilenberg G. Babesia - A historical overview. Vet Parasitol. 2006;138:3–10.
Criado-Fornelio A, Buling A, Pingret JL, Etievant M, Boucraut-Baralon C, Alongi A, et al. Hemoprotozoa of domestic animals in France: prevalence and molecular characterization. Vet Parasitol. 2009;159:73–6.
Homer MJ, Aguilar-Delfin I, Telford SR, Krause PJ, Persing DH. Babesiosis. Clin Microbiol Rev. 2002;13:451–69.
Yabsley MJ, Work TM, Rameyer RA. Molecular phylogeny of Babesia poelea from brown boobies (Sula leucogaster) from Johnston Atoll, central Pacific. J Parasitol. 2006;92:423–5.
Vannier E, Gewurz BE, Krause PJ. Human babesiosis. Infect Dis Clin North Am. 2008;22:469–88.
Schnittger L, Rodriguez AE, Florin-Christensen M, Morrison DA. Babesia: A world emerging. Infect Genet Evol. 2012;12:1788–809.
Vannier E, Krause PJ. Update on babesiosis. Interdiscip Perspect Infect Dis. 2009;984568. doi:10.1155/2009/984568.
Bock R, Jackson L, de Vos A, Jorgensen W. Babesiosis of cattle. Parasitology. 2004;129:S247–69.
Ranjbar-Bahadori S, Eckert B, Omidian Z, Shirazi NS, Shayan P. Babesia ovis as the main causative agent of sheep babesiosis in Iran. Parasitol Res. 2012;110:1531–6.
Guidi E, Pradier S, Lebert I, Leblond A. Piroplasmosis in an endemic area: analysis of the risk factors and their implications in the control of Theileriosis and Babesiosis in horses. Parasitol Res. 2014; doi:10.1007/s0043601441619.
Schoeman JP. Canine babesiosis. Onderstepoort J Vet Res. 2009;76:59–66.
Wang C, Ahluwalia SK, Li Y, Gao D, Poudel A, Chowdhury E, et al. Frequency and therapy monitoring of canine Babesia spp. infection by high-resolution melting curve quantitative FRET-PCR. Vet Parasitol. 2010;168:11–8.
Kamani J, Baneth G, Mumcuoglu KY, Waziri NE, Eyal O, Guthmann Y, et al. Molecular detection and characterization of tick-borne pathogens in dogs and ticks from Nigeria. PLoS Negl Trop Dis. 2013;7:e2108.
Ibrahim HM, Adjou Moumouni PF, Mohammed-Geba K, Sheir SK, Hashem IS, Cao S, et al. Molecular and serological prevalence of Babesia bigemina and Babesia bovis in cattle and water buffalos under small-scale dairy farming in Beheira and Faiyum Provinces, Egypt. Vet Parasitol. 2013;198:187–92.
Liu J, Guan G, Liu A, Li Y, Yin H, Luo J. A PCR method targeting internal transcribed spacers: the simultaneous detection of Babesia bigemina and Babesia bovis in cattle. Acta Parasitol. 2014;59:132–8.
Aktaş M, Altay K, Dumanli N. Development of a polymerase chain reaction method for diagnosis of Babesia ovis infection in sheep and goats. Vet Parasitol. 2005;133:277–81.
Chi Q, Liu Z, Li Y, Yang J, Chen Z, Yue C, et al. Development of a real-time PCR assay for detection and quantification of Anaplasma ovis infection. Transbound Emerg Dis. 2013;60 Suppl 2:119–24.
Kelly PJ, Lucas H, Yowell C, Beati L, Dame J, Urdaz-Rodriguez J, et al. Ehrlichia ruminantium in Amblyomma variegatum and domestic ruminants in the Caribbean. J Med Entomol. 2011;48:485–8.
Wei L, Kelly P, Zhang J, Yang Y, Zheng X, Tao J, et al. Use of a universal hydroxymethylbilane synthase (HMBS)-based PCR as an endogenous internal control and to enable typing of mammalian DNAs. Appl Microbiol Biotechnol. 2014;98:5579–87.
Kelly PJ, Xu C, Lucas H, Loftis A, Abete J, Zeoli F, et al. Ehrlichiosis, Babesiosis, Anaplasmosis and Hepatozoonosis in dogs from St. Kitts, West Indies. PLoS One. 2013;8:e53450.
Wang C, Gao D, Vaglenov A, Kaltenboeck B. One-step real-time duplex reverse transcription PCRs simultaneously quantify analyte and housekeeping gene mRNAs. Biotechniques. 2004;36:508–16. 518–519.
Yang Y, Mao Y, Kelly P, Yang Z, Luan L, Zhang J, et al. A pan-Theileria FRET-qPCR survey for Theileria spp in ruminants from nine provinces of China. Parasit Vectors. 2014;7:413.
Stackkebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–5.
Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013; doi:10.1186/147120151460.
Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:1825.
Garris GI, Scotland K. Ticks on livestock in St. Lucia. Vet Parasitol. 1985;18:367–73.
Camus E, Barre N. Vector situation of tick-borne diseases in the Caribbean islands. Vet Parasitol. 1995;57:167–76.
Hadrill DJ, Boid R, Jones TW, Bell-Sakyi L. Bovine babesiosis on Nevis-implications for tick control. Vet Record. 1990;126:403–4.
Alonso M, Camus E, Rodriguez Diego J, Bertaudière L, Tatareau JC, Liabeuf JM. Current status of bovine haemoparasitic diseases in Martinique (French West Indies). Rev Elev Med Vet Pays Trop. 1992;45:9–14.
Camus E, Montenegro-James S. Bovine anaplasmosis and babesiosis in the Lesser Antilles: Risk assessment of unstable situation. Vet Res. 1994;25:313–7.
Rampersad J, Cesar E, Campbell MD, Samlal M, Ammons D. A field evaluation of PCR for the routine detection of Babesia equi in horses. Vet Parasitol. 2003;114:81–7.
Asgarali Z, Coombs DK, Mohammed F, Campbell MD, Caesar E. A serological study of Babesia caballi and Theileria equi in Thoroughbreds in Trinidad. Vet Parasitol. 2007;144:167–71.
Urdaz-Rodríguez JH, Fosgate GT, Waghela SD, Alleman AR, Rae DO, Donovan GA, et al. Seroprevalence estimation and management factors associated with high herd seropositivity for Babesia bovis in commercial dairy farms of Puerto Rico. Trop Anim Health Prod. 2009;41:1465–73.
Mulugeta Y, Yacob HT, Ashenafi H. Ectoparasites of small ruminants in three selected agro-ecological sites of Tigray Region, Ethiopia. Trop Anim Health Prod. 2010;42:1219–24.
Friedhoff KT, Tenter AM, Müller I. Haemoparasites of equines: impact on international trade of horses. Rev Sci Tech. 1990;9:1187–94.
Corn JL, Berger P, Mertins JW. Surveys for ectoparasites on wildlife associated with Amblyomma variegatum (Acari: Ixodidae)-infested livestock in St. Croix, U.S. Virgin Islands. J Med Entomol. 2009;46:1483–9.
Goff WL, Jessup DA, Waldrup KA, Thomford JW, Conrad PA, Boyce WM, et al. The isolation and partial characterization of a Babesia sp. from desert bighorn sheep (Ovis canadensis nelsoni). J Eukaryot Microbiol. 1993;40:237–43.
Yabsley MJ, McKibben J, Macpherson CN, Cattan PF, Cherry NA, Hegarty BC, et al. Prevalence of Ehrlichia canis, Anaplasma platys, Babesiacanis vogeli, Hepatozoon canis, Bartonella vinsonii berkhoffii, and Rickettsia spp. in dogs from Grenada. Vet Parasitol. 2008;151:279–85.
Loftis A, Kelly PJ, Freeman M. Beeler, Wang C. Tick-borne pathogens and disease of dogs in St. Kitts, West Indies. Vet Parasitol. 2013;196:44–9.
Simking P, Wongnakphet S, Stich RW, Jittapalapong S. Detection of Babesia vogeli in stray cats of metropolitan Bangkok. Thailand Vet Parasitol. 2010;173:70–5.
Kelly P, Marabini L, Dutlow K, Zhang J, Loftis A, Wang C. Molecular detection of tick-borne pathogens in captive wild felids, Zimbabwe. Parasit Vectors. 2014;7:514.
Bouattour A, Darghouth MA, Daoud A. Distribution and ecology of ticks (Acari: Ixodidae) infesting livestock in Tunisia: an overview of eighth years field collections. Parassitologia. 1999;41(Suppl):5–10.
Torina A, Khoury C, Caracappa S, Maroli M. Ticks infesting livestock on farms in Western Sicily, Italy. Exp Appl Acarol. 2006;38:75–86.
Harrus S, Perlman-Avrahami A, Mumcuoglu KY, Morick D, Eyal O, Baneth G. Molecular detection of Ehrlichia canis, Anaplasma bovis, Anaplasma platys, Candidatus Midichloria mitochondrii and Babesia canis vogeli in ticks from Israel. Clin Microbiol Infect. 2011;17:459–63.
Dantas-Torres F, Latrofa MS, Annoscia G, Giannelli A, Parisi A, Otranto D. Morphological and genetic diversity of Rhipicephalus sanguineus sensu lato from the New and Old Worlds. Parasit Vectors. 2013;6:213.
Wei L, Kelly P, Ackerson K, Zhang J, El-Mahallawy HS, Kaltenboeck B, et al. First report of Babesia gibsoni in Central America and survey for vector-borne infections in dogs from Nicaragua. Parasit Vector. 2014;25:126.
Orkun Ö, Karaer Z, Çakmak A, Nalbantoğlu S. Identification of tick-borne pathogens in ticks feeding on humans in Turkey. PLoS Negl Trop Dis. 2014;8:e3067.
Acknowledgments
This project was supported by the Ross University School of Veterinary Medicine, the Priority Academic Program Development of Jiangsu Higher Education Institutions, Yangzhou, Jiangsu, P. R. China and grants from the National Natural Science Foundation of China (NO: 31272575) and the United States Department of Agriculture through its program for Tropical and Subtropical Agricultural Research (T-STAR) (Grant 2006 34135 6930).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
WC, LJ and ZJ designed the study and performed the statistical analysis. LJ and XC participated in the experiments. WC and KP wrote the manuscript. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, J., Kelly, P., Zhang, J. et al. Development of a pan-Babesia FRET-qPCR and a survey of livestock from five Caribbean islands. BMC Vet Res 11, 246 (2015). https://doi.org/10.1186/s12917-015-0560-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-015-0560-0