Abstract
Background
Congenital cataract (CC) is a significant cause of lifelong visual loss, and its genetic diagnosis is challenging due to marked genetic heterogeneity. The purpose of this article is to report the genetic findings in sporadic and familial CC patients.
Methods
Patients (n = 53) who were clinically diagnosed with CC and their parents were recruited. Blood samples were collected in our hospital. Mutations were detected by panel-based next-generation DNA sequencing (NGS) targeting 792 genes frequently involved in common inherited eye diseases.
Results
We identified variants in 10/37 cases (27.02%) of sporadic CC and 14/16 cases (87.5%) of familial CC, which indicated a significant difference (P = 0.000). Of the 13 variants identified in sporadic cases, nine were previously reported mutations, and three were novel mutations, including one de novo mutation (CRYBB2 c.487C > T). The most frequent variants in our cohort were in crystallins and cytoskeletal genes (5/27, 18.52%), followed by proteins associated with X-linked syndromic conditions (14.81%) and transcriptional factors (11.11%). Additional information on the possibility of complications with inherited ocular or systemic diseases other than CC was provided in 17/27 (62.96%) variants.
Conclusions
These results contribute to expanding the mutation spectrum and frequency of genes responsible for CC. Targeted NGS in CC provided significant diagnostic information and enabled more accurate genetic counselling. This study reports the different distributions of mutation genes in familial and sporadic CC cases.
Similar content being viewed by others
Background
Congenital cataracts (CCs) are now the most common avoidable cause of childhood blindness worldwide, accounting for 10–35% of such cases, with an estimated incidence of 0.63–9.14/10,000 births [1,2,3,4]. Management is often difficult due to the risk of amblyopia in the developing visual system and complications of glaucoma, posterior synechia or visual axis opacification, which require additional surgery [5]. CCs occur due to the disruption of the lens microarchitecture or the protein function in the lens [6]. Except for a very few infectious cases, only one-third of CC cases have a positive family history [7], with the other two-thirds having an unknown aetiology [8]. Therefore, a significant proportion are sporadic cases in which it is not known whether there is an underlying genetic cause for the lens abnormality.
Thus far, approximately 350 genes have been reported to be associated with CC (Cat-Map; http://cat-map.wustl.edu/); these include mutations in crystallins and gap junction, membrane transport and channel, and cytoskeletal proteins and growth and transcription factors [9]. Locating and identifying the involved genes and mutations are essential to gaining an understanding of the molecular defects and pathophysiologic characteristics underlying inherited CC.
A conventional approach to identifying mutations in CC is usually performed by Sanger sequencing only in familial cases and is time-consuming and costly, with a detection rate of 30–50% in apparent autosomal dominant cases [10, 11]. Due to marked genetic and phenotypic heterogeneity, determining the precise genetic cause of CC and establishing a robust genotype-phenotype correlation is challenging. Next-generation DNA sequencing (NGS) is increasingly powerful as a diagnostic tool and offers speed, precision, and cost-effectiveness for heterogeneous conditions [12]. This has been demonstrated in studies to determine the cause of other heterogeneous inherited eye diseases, such as congenital macular dystrophy and retinal pigmentosa [13,14,15,16]. Recent studies have also shown that NGS allows the efficient identification of genetic causes of CC in the majority of cases, thereby improving its diagnosis and clarifying inheritance patterns [17,18,19] while guiding genetic counselling and increasing prognostic accuracy.
In this study, we applied targeted NGS in 792 genes involved in common inherited eye diseases to detect causal mutations in a relatively large series of CCs, including a high proportion of sporadic cases, and report the different distributions of mutated genes in sporadic versus familial CC cases (sCC VS fCC), while broadening the mutation spectrum and frequency of genes responsible for CC.
Methods
Ethical statement
All participants (parents on behalf of their children) provided written informed consent forms for both genetic counselling and molecular genetic testing prior to enrolment. The study was approved by the Ethics Committee of the Eye and ENT Hospital of Fudan University. All research was conducted in accordance with the Declaration of Helsinki.
Clinical evaluations and sample collection
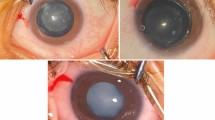
Patients who were clinically diagnosed with CC from June 2018 to May 2019 were recruited. All patients underwent a detailed ophthalmic examination, including slit-lamp examination, B ultrasound, intraocular pressure measurement, and ultrasonic A-scan, as mentioned in our previous study [20]. Visual acuity (VA) was recorded in all patients who were able to cooperate. Patients diagnosed with monocular CC additionally underwent post-eyeball colour Doppler ultrasound to help in the differential diagnosis of persistent hyperplastic primary vitreous (PHPV). Children younger than 3 years old were examined under sedation with chloral hydrate. The lens phenotypes of patients and their parents were carefully recorded in all families and included childbirth history, medical history, family history and a detailed history of the gestation period, including high fever, rubella virus [RV] TORCHES ([Toxoplasma gondii; T. gondii], cytomegalovirus [CMV], herpes simplex virus [HSV], syphilis [caused by Treponema pallidum]), tuberculosis infection, exposure to radiation, and drug intake. Additional systemic problems were also recorded in patients and included serum biochemical tests for levels of blood glucose, calcium and phosphorous as well as urine tests. The probands for whom at least one immediate family member had a history of CC were defined as familial cases. Those who had no family history and had been excluded from infection factors were classified as sporadic cases. Blood samples were collected in children while under general anaesthesia during eye surgery and from their biological parents (Trio sequencing) in our hospital. In familial cases, blood samples of other available affected relatives were also collected.
Next-generation sequencing
Genomic DNA was extracted from peripheral blood samples using standard methods. Panel-based NGS was performed on all subjects in this study. We designed the Target_Eye_792_V2 chip with exon-capture and untranslated regions (UTRs) of 792 genes most frequently involved in common inherited eye diseases (Additional file 1, Table S1), which were produced by BGI-Shenzhen, Guangdong, China as previously reported [21]. Then, DNA fragments were sequenced by an Illumina HiSeq 2000 platform (Illumina, Inc., San Diego, CA, United States). The following databases were then used to annotate all identified variants with a minor allele frequency (MAF) > 0.1% to eliminate benign variants as previously described [22]: dbSNP1371 (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/database/snp137.txt.gz), HapMap Project (ftp://ftp.ncbi.nlm.nih.gov/hapmap), 1000 Genomes Project (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp), YH database (http://yh.genomics.org.cn/), and Exome Variant Server (http://evs.gs.washington.edu/EVS/). Subsequently, variant prioritization was performed to combine the total depth, quality score, MAF, potential deleterious effect and existence of mutation reports in common databases such as the Human Gene Mutation Database (HGMD), ClinVar or Online Mendelian Inheritance in Man (OMIM). Finally, variants were classified as pathogenic, likely pathogenic, uncertain significance according to the American College of Medical Genetics (ACMG) and genomics guidelines. Sanger sequencing was performed to confirm the candidate variants.
Results
Participant characteristics
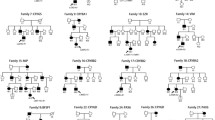
A total of 152 subjects of 53 families were recruited in this study, including 16 familial cases (49 subjects in total) and 37 sporadic cases (106 subjects in total). Parental samples in one familial case and six sporadic cases were not completely obtained for some reasons beyond control. All the familial cases had at least one affected parent (11 mothers and 5 fathers). In addition, the available affected immediate relatives, the brother, the paternal grandfather and the maternal grandmother in three familial cases participated in the test. The mean ages of the 53 children and their mothers and fathers were 3.0 [1.50–6.00], 30.72 ± 5.02, and 32.65 ± 5.19 years old, respectively. There were more binocular cases than monocular cases and more male than female cases. More detailed information is presented in Table 1. No significant differences were found between sCC and fCC in the mean ages of children and the parents or other constituent ratios (P values are presented in Table 1).
Variants identified
A total of 27 variants were found in 24 of the 53 patients with CC in our cohort, yielding a total detection rate of 45.30%. We identified variants in 10/37 (27.03%) sCC and 14/16 (87.5%) fCC cases, indicating a significant difference (P = 0.000, Table 1). The detection rate was lower in monocular cases (4/12, 33.33%) than in binocular cases (20/41,48.80%), but the difference was not significant (P = 0.512).
The variants detected are presented in Table 2 and Table 3. According to the ACMG mutation guidelines, 17 of 27 variants were classified as pathogenetic, five were likely pathogenic, and seven were uncertain significance (VUS).
We identified three novel likely cataractous causative mutations in sCC in CRYBB2 and NHS (*2), one of which was a de novo mutation in CRYBB2 c.487C > T (p. Gln163*|p. Q163*). Eight of the 27 variants detected in our cohort were previously reported pathogenic gene mutations in CC, including loci in CRYGC, CRYGD(*2), CRYAA, CRYBA1, and GJA8 and adjacent loci in CRYGC and PAX6. Another 16 variants involved in additional ocular or systemic diseases that had been reported or included by HGMD or ClinVar were also identified, including CYP27A1, OPA3, JAG1, BEST1, BMP4, CYP1B1, and TSPAN12 (see Tables 2 and 3, Note column).
In terms of gene function, genes encoding crystallins were the most frequently identified in our cohort, accounting for 7/27 (25.93%) of the cases, followed by cytoskeletal proteins (18.52%), X-linked syndromic proteins (14.81%) and transcriptional factors (11.11%).
Differential distribution of mutational genes
A comparison of the distributions of mutational genes between fCC and sCC showed that variants in crystallins accounted for the highest proportion (37.50%) in fCC cases but only 9.00% in sCC cases (Fig. 1). The sporadic cases mainly consisted of X-linked syndromic proteins and structural protein genes, including transmembrane and collagen-associated proteins.
Discussion
Approximately 70% of CC cases may occur alone, and 15% of such cases may be accompanied by other ocular abnormalities, such as microphthalmia, aniridia, or retinal degeneration. In another 15% of cases, cataracts are one part of a multisystem genetic disorder [47]. To obtain clues related to the noncataractous phenotype, we designed a panel with exon-capture and NGS targeting of the 792 genes most frequently involved in common inherited eye diseases. Compared to related previous studies, our study included the largest numbers of patients and targeted genes. We achieved detection rates in familial and sporadic cases similar to those in a recent study [37]. Although the overall detection rate (45.3%) in our cohort was apparently lower than that in the other studies listed in Table 4, these rates are not comparable due to differences in the proportions of participants. Most of the studies [17,18,19] included only binocular cataracts, whereas we enrolled many monocular cases. Regarding the distribution of genes, our result was slightly different from those reported previously. Li et al. [37] reported that variants in the crystallin genes were the most frequent mutations found in their study, whether in familial or sporadic cases. We found that variants in crystallins accounted for a similar proportion of fCC cases but only 1 sCC case (Fig. 1). X-linked syndromic proteins and structural protein genes, such as transmembrane and collagen-associated proteins, accounted for most of the sCCs in our study.
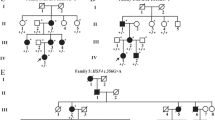
In our study, approximately 17/27 (62.96%) variants provided clues regarding the possibility of complication with inherited ocular or systemic diseases other than CC. Among these, nine identified loci provided additional ophthalmological diagnostic information. For instance, OPA3 mutations are associated with optic atrophy [24], BEST1 mutations are associated with best vitelliform macular dystrophy (BEST) [27,28,29,30], TSPAN12 mutations are associated with familial exudative vitreoretinopathy (FEVR) [35], PAX6 mutation are associated with aniridia and Peter’s anomaly [48], and CYP1B1 mutations are associated with glaucoma [45]. In addition, we also identified a monoallelic mutation in BMP4, which has been mostly associated with microphthalmia [40] or facial clefts [49]. Eight variants were associated with systemic syndrome. WFS1 is the most common causative gene in Wolfram-like syndrome, a rare autosomal dominant disease characterized by congenital progressive hearing loss, diabetes mellitus, and optic atrophy [50]. COL4A5 is one of the causative genes in Alport Syndrome, a genetic condition characterized by progressive loss of kidney function, hearing, and eye abnormalities, including misshapen lenses and abnormal retina [34]. JAG1 has been associated with Alagille syndrome, which involves cholestasis, cardiac defects, ocular abnormalities, skeletal abnormalities and characteristic faces. Loss-of-function mutations in the BCOR gene have been identified in individuals with oculo-facio-cardio-dental syndrome (OFCD), which includes microcornea, CC, and facial, cardiac, and dental abnormalities [38]. Mutations in the FBN1 (fibrillin-1) gene may be diagnostic of Marfan syndrome [46]. NHS mutations have been identified in patients with Nance-Horan syndrome (NHS), an X-linked developmental disorder characterized by CC, dental anomalies, facial dysmorphism and, in some cases, mental retardation [51]. Clinically, a new diagnosis was made after surgery and with reference to genetic testing in at least two patients in our cohort. One of the sporadic cases (ID 10 in Table 2) presented some retinal abnormalities during operations after the removal of cataracts in both eyes, including settled subretinal exudates and dragging of the optic disc. Combined with this clinical manifestation, we have clarified the diagnosis of FEVR with regard for the TSPAN12 mutation, which is a pathogenic gene known to indicate FEVR. We also observed dental, facial and mental anomalies and made a new diagnosis of NHS at 2 years after the first CC operation was performed in one of the sporadic cases with an identified NHS mutation (ID 6 in Table 2). However, whether other variants are associated with a noncataractous phenotype is difficult to confirm. For example, in family 3 (Table 2), we cannot clearly ascribe one of these variants, OPA3 or JAG1 to a cataractogenic effect. It is possible that one or more gene mutations cause multiple eye abnormalities at the same time, and cataracts are only one of the first manifestations found in the clinic. During the follow-up period after cataract surgery, we will pay more attention to whether the child tends to experience optic atrophy and will give suggestions for monitoring liver and cardiac function. The relationships between complicated phenotypes and mutations in ocular genes are not explicit. Thus, more cases should be included, and more experiments should be performed to verify these connections.
It is worth emphasizing that those identified variants in non-classical cataract genes may not be initially ascribed to a cataractogenic effect. They might indicate other inherited eye disorders or syndromes, in which a cataractous phenotype may not be presented in every carrier. Another possibility was that the exact cataractous causative genes are located in regions that have not yet been detected, or even that the cataractous phenotype was not caused by genetic factors at all or may involve epigenetic factors.
The phenomenon in which identified heterozygous variants are also present in unaffected parents in sporadic cases (Table 2) might be explained by incomplete and variable penetrance; the underlying mechanisms of this phenomenon remain largely unknown. A recent study also provides support showing that variants associated with inherited eye disorders are frequently encountered in unaffected individuals and that one in six genes implicated in inherited eye disorders are potentially associated with variable penetrance [52]. The number of variants and genes that do not segregate (Table 2) is relatively high in our study. Some of these genes, such as BEST1 and WFS1, were shown to exhibit variable penetrance in a previous study [50]. Incomplete penetrance of the remaining genes might not be supported by sufficient evidence, or these genes might not be the causative genes. This phenomenon might also be due to the limited number of samples detected. In a future study, we will continue to expand the sample size, collect more samples of family members, and improve the history tracking. We believe that the proportion of this phenomenon will be significantly reduced.
This study emphasizes the power and necessity for trio NGS analyses of CC families. By identifying pathogenic heterozygous and homozygous mutations, de novo mutations, and parental mosaicism, such analyses may reveal a new pattern of inheritance in CC with significance not limited to the affected child. However, trio NGS can reveal numerous VUS, for which functional validation is mandatory, although it is still a challenge. Furthermore, future research is required to determine the clinical significance of non-Mendelian inheritance, the intricate mutual effect between genetic predispositions and environmental factors, and interactions between genetic and epigenetic. These studies will provide important insights into the pathogenesis and the complex genotype-to-phenotype association of CC. In the future, these results may also lead to the development of novel gene therapies for some types of congenital cataracts, similar to other inherited eye diseases.
A limitation of this study is that samples in which no mutations were identified could be further submitted to whole-genome sequencing but rarely are because it is challenging to obtain a sufficient amount of blood from infants and young children to meet experimental needs.
Conclusions
In conclusion, our study highlights the benefits of an NGS approach combined with the analysis of a large targeted group of genes in a setting of genetically heterogeneous CC patients. Our findings provide significant diagnostic information and enable more accurate genetic counselling. Our results expand what we know about the mutation spectrum and frequencies of genes responsible for CC as well as the different distributions of genes mutated in familial and sporadic cases in the Chinese population.
Availability of data and materials
The raw sequencing datasets generated during the current study are not publicly available due to legal requirements in accordance with The Regulations of People’s Republic of China on Administration of Human Genetic Resources. However, original analyzed data of the sequencing datasets (excluding patient information) are available from the corresponding author on reasonable request.
Abbreviations
- CC:
-
Congenital cataract
- sCC:
-
Sporadic congenital cataract
- fCC:
-
Familial congenital cataract
- NGS:
-
Next-generation DNA sequencing
- HGMD:
-
Human Gene Mutation Database
- ACMG:
-
American College of Medical Genetics
- VUS:
-
Uncertain significance
- FEVR:
-
Familial exudative vitreoretinopathy
- BEST:
-
Best vitelliform macular dystrophy
- NHS:
-
Nance-Horan syndrome
References
Daien V, Le Pape A, Heve D, Villain M, Gignac DB, Safety CE. Incidence and characteristics of congenital cataract surgery in France from 2010 to 2012: the EPISAFE program. Ophthalmic Res. 2017;58(2):114–6.
Wu XH, Long EP, Lin HT, Liu YZ. Global prevalence and epidemiological characteristics of congenital cataract: a systematic review and meta-analysis. Lancet. 2016;388:55.
Sheeladevi S, Lawrenson JG, Fielder AR, Suttle CM. Global prevalence of childhood cataract: a systematic review. Eye (Lond). 2016;30(9):1160–9.
Solebo AL, Teoh L, Rahi J. Epidemiology of blindness in children. Arch Dis Child. 2017;102(9):853–7.
Vasavada AR, Vasavada V, Shah SK, Praveen MR, Vasavada VA, Trivedi RH, et al. Five-year postoperative outcomes of bilateral Aphakia and Pseudophakia in children up to 2 years of age: a randomized clinical trial. Am J Ophthalmol. 2018;193:33–44.
Shiels A, Hejtmancik JF. Mutations and mechanisms in congenital and age-related cataracts. Exp Eye Res. 2017;156:95–102.
Wirth MG, Russell-Eggitt IM, Craig JE, Elder JE, Mackey DA. Aetiology of congenital and paediatric cataract in an Australian population. Brit J Ophthalmol. 2002;86(7):782–6.
Haargaard B, Wohlfahrt J, Fledelius HC, Rosenberg T, Melbye M. A nationwide Danish study of 1027 cases of congenital/infantile cataracts - etiological and clinical classifications. Ophthalmology. 2004;111(12):2292–8.
Shiels A, Bennett TM, Hejtmancik JF. Cat-map: putting cataract on the map. Mol Vis. 2010;16:2007–15.
Sun WM, Xiao XS, Li SQ, Guo XM, Zhang QJ. Mutational screening of six genes in Chinese patients with congenital cataract and microcornea. Mol Vis. 2011;17(168–69):1508–13.
Hansen L, Yao WL, Eiberg H, Kjaer KW, Baggesen K, Hejtmancik JF, et al. Genetic heterogeneity in microcornea-cataract: five novel mutations in CRYAA, CRYGD, and GJA8. Invest Ophthalmol Vis Sci. 2007;48(9):3937–44.
Shendure J, Ji HL. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–45.
Simpson DA, Clark GR, Alexander S, Silvestri G, Willoughby CE. Molecular diagnosis for heterogeneous genetic diseases with targeted high-throughput DNA sequencing applied to retinitis pigmentosa. J Med Genet. 2011;48(3):145–51.
Coppieters F, De Wilde B, Lefever S, De Meester E, De Rocker N, Van Cauwenbergh C, et al. Massively parallel sequencing for early molecular diagnosis in Leber congenital amaurosis. Genet Med. 2012;14(6):576–85.
Shanks ME, Downes SM, Copley RR, Lise S, Broxholme J, Hudspith KAZ, et al. Next-generation sequencing (NGS) as a diagnostic tool for retinal degeneration reveals a much higher detection rate in early-onset disease. Eur J Hum Genet. 2013;21(3):274–80.
Valencia CA, Ankala A, Rhodenizer D, Bhide S, Littlejohn MR, Keong LM, et al. Comprehensive mutation analysis for congenital muscular dystrophy: a clinical PCR-based enrichment and next-generation sequencing panelPLoS One. 2013;8(1):e53083.
Gillespie RL, O'Sullivan J, Ashworth J, Bhaskar S, Williams S, Biswas S, et al. Personalized diagnosis and management of congenital cataract by next-generation sequencing. Ophthalmology. 2014;121(11):2124–U302.
Zhai Y, Li JY, Yu WS, Zhu S, Yu YH, Wu MH, et al. Targeted exome sequencing of congenital cataracts related genes: broadening the mutation spectrum and genotype-phenotype correlations in 27 Chinese Han families. Sci Rep-Uk. 2017;7:1.
Ma AS, Grigg JR, Ho G, Prokudin I, Farnsworth E, Holman K, et al. Sporadic and familial congenital cataracts: mutational spectrum and new diagnoses using next-generation sequencing. Hum Mutat. 2016;37(4):371–84.
Liu X, Zheng TY, Zhou XT, Lu Y, Zhou P, Fan F, et al. Comparison between limbal and pars plana approaches using microincision vitrectomy for removal of congenital cataracts with primary intraocular lens implantation. J Ophthalmol. 2016;2016:8951053.
Hu FY, Li JK, Gao FJ, Qi YH, Xu P, Zhang YJ, et al. ABCA4 gene screening in a Chinese cohort with Stargardt disease: identification of 37 novel variants. Front Genet. 2019;10:773.
Guo J, Gao F, Tang W, Qi Y, Xuan Y, Liu W, et al. Novel Best1 mutations detected by next-generation sequencing in a Chinese population with vitelliform macular dystrophy. Retina. 2019;39(8):1613–22.
Khan AO, Aldahmesh MA, Alkuraya FS. Phenotypes of Recessive Pediatric Cataract in a Cohort of Children with Identified Homozygous Gene Mutations (An American Ophthalmological Society Thesis). Trans Am Ophthalmol Soc. 2015;113:T7.
Chen JQ, Xu K, Zhang XH, Jiang F, Liu LJ, Dong B, et al. Mutation screening of mitochondrial DNA as well as OPA1 and OPA3 in a Chinese cohort with suspected hereditary optic atrophy. Invest Ophthalmol Vis Sci. 2014;55(10):6987.
Qian YY, Xiao DY, Guo X, Chen HB, Hao LL, Ma XJ, et al. Multiple gene variations contributed to congenital heart disease via GATA family transcriptional regulation. J Transl Med. 2017;15.
Warthen DM, Moore EC, Katnath BM, Morrissette JJD, Sanchez P, Piccoli DA, et al. Jagged1 (JAG1) mutations in alagille syndrome: Increasing the mutation detection rate. Human Mutation. 2006;27(5):436-43.
Katagiri S, Hayashi T, Ohkuma Y, Sekiryu T, Takeuchi T, Gekka T, et al. Mutation analysis of BEST1 in Japanese patients with Best's vitelliform macular dystrophy. Brit J Ophthalmol. 2015;99(11):1577–82.
Tian L, Sun TY, Xu K, Zhang XH, Peng XY, Li Y. Screening of BEST1 gene in a Chinese cohort with BEST vitelliform macular dystrophy or autosomal recessive bestrophinopathy. Invest Ophthalmol Vis Sci. 2017;58(9):3366–75.
Nakanishi A, Ueno S, Hayashi T, Katagiri S, Kominami T, Ito Y, et al. Clinical and genetic findings of autosomal recessive bestrophinopathy in Japanese cohort. Am J Ophthalmol. 2016;168:86–94.
Borman AD, Davidson AE, O'Sullivan J, Thompson DA, Robson AG, De Baere E, et al. Childhood-onset autosomal recessive bestrophinopathy. Arch Ophthalmol-Chic. 2011;129(8):1088–93.
Sloan-Heggen CM, Bierer AO, Shearer AE, Kolbe DL, Nishimura CJ, Frees KL, et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Human Genetics. 2016;135(4):441-50.
Prochazkova D, Hruba Z, Konecna P, Skotakova J, Fajkusova L. A p.(Glu809Lys) Mutation in the WFS1 Gene Associated with Wolfram-like Syndrome: A Case Report. J Clin Res Pediatr Endocrinol. 2016;8(4):482-3.
Qing J, Yan D, Zhou Y, Liu Q, Wu WJ, Xiao Z, et al. Whole-Exome Sequencing to Decipher the Genetic Heterogeneity of Hearing Loss in a Chinese Family with Deaf by Deaf Mating. PloS one. 2014;9(10).
Miyagawa M, Naito T, Nishio S, Kamatani N, Usami S. Targeted exon sequencing successfully discovers rare causative genes and clarifies the molecular epidemiology of japanese deafness patients. PLoS One. 2013;8(8):e71381.
Kondo H, Kusaka S, Yoshinaga A, Uchio E, Tawara A, Hayashi K, et al. Mutations in the TSPAN12 gene in Japanese patients with familial exudative vitreoretinopathy. Am J Ophthalmol. 2011;151(6):1095-100 e1.
Mackay DS, Bennett TM, Culican SM, Shiels A. Exome sequencing identifies novel and recurrent mutations in GJA8 and CRYGD associated with inherited cataract. Human Genomics. 2014;8.
Li JY, Leng YJ, Han SR, Yan LL, Lu CX, Luo Y, et al. Clinical and genetic characteristics of Chinese patients with familial or sporadic pediatric cataract. Orphanet J Rare Dis. 2018;13:94.
Danda S, van Rahden VA, John D, Paul P, Raju R, Koshy S, et al. Evidence of germline mosaicism for a novel BCOR mutation in two Indian sisters with Oculo-Facio-Cardio-Dental syndrome. Mol Syndromol. 2014;5(5):251–6.
Sun ZX, Zhou Q, Li HJ, Yang LZ, Wu SJ, Sui RF. Mutations in crystallin genes result in congenital cataract associated with other ocular abnormalities. Molecular Vision. 2017;23:977-86.
Zhang X, Li S, Xiao X, Jia X, Wang P, Shen H, et al. Mutational screening of 10 genes in Chinese patients with microphthalmia and/or coloboma. Mol Vis. 2009;15:2911–8.
Zhong ZL, Wu ZH, Han LY, Chen JJ. Novel mutations in CRYGC are associated with congenital cataracts in Chinese families. Sci Rep-Uk. 2017;7.
Sarkar D, Ray K, Sengupta M. Structure-Function Correlation Analysis of Connexin50 Missense Mutations Causing Congenital Cataract: Electrostatic Potential Alteration Could Determine Intracellular Trafficking Fate of Mutants. Biomed Res Int. 2014.
Azuma N, Yamada M. Missense mutation at the C terminus of the PAX6 gene in ocular anterior segment anomalies. Invest Ophthalmol Vis Sci. 1998;39(5):828–30.
Azuma N, Hotta Y, Tanaka H, Yamada M. Missense mutations in the PAX6 gene in aniridia. Invest Ophthalmol Vis Sci. 1998;39(13):2524–8.
Gong B, Qu C, Li XL, Shi Y, Lin Y, Zhou Y, et al. Mutation spectrum of CYP1B1 in Chinese patients with primary open-angle glaucoma. Brit J Ophthalmol. 2015;99(3):425–30.
Comeglio P, Johnson P, Arno G, Brice G, Evans A, Aragon-Martin J, et al. The importance of mutation detection in Marfan syndrome and Marfanrelated disorders: report of 193 FBN1 mutations. Hum Mutat. 2007;28(9):928.
Hejtmancik JF. Congenital cataracts and their molecular genetics. Seminars in cell & developmental biology. 2008;19(2):134–49.
Davis LK, Meyer KJ, Rudd DS, Librant AL, Epping EA, Sheffield VC, et al. Pax6 3 ' deletion results in aniridia, autism and mental retardation. Hum Genet. 2008;123(4):371–8.
Suzuki S, Marazita ML, Cooper ME, Miwa N, Hing A, Jugessur A, et al. Mutations in BMP4 are associated with subepithelial, microform, and overt cleft lip. American journal of human genetics. 2009;84(3):406–11.
De Franco E, Flanagan SE, Yagi T, Abreu D, Mahadevan J, Johnson MB, et al. Dominant ER Stress-Inducing WFS1 Mutations Underlie a Genetic Syndrome of Neonatal/Infancy-Onset Diabetes, Congenital Sensorineural Deafness, and Congenital Cataracts. Diabetes. 2017;66(7):2044–53.
Coccia M, Brooks SP, Webb TR, Christodoulou K, Wozniak IO, Murday V, et al. X-linked cataract and Nance-Horan syndrome are allelic disorders. Hum Mol Genet. 2009;18(14):2643–55.
Green DJ, Sallah SR, Ellingford JM, Lovell SC, Sergouniotis PI. Variability in Gene Expression is Associated with Incomplete Penetrance in Inherited Eye Disorders. Genes-Basel. 2020;11(2).
Acknowledgements
We thank our collaborator BGI (Shenzhen, China) for providing the sequencing platforms. We are grateful to the members of the families for their participation in the study. We would like to thank all the residents and clinicians for their efforts during sample collection. We are also grateful to Fangyuan Hu for the constructive suggestions during the revision process.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant No 81870645) and the National Natural Youth Fund (Grant No 81900839), which provided support for the collection of the samples and the writing of the manuscript. This work was carried out in collaboration with BIG (Shenzhen, China), which completed the sequencing.
Author information
Authors and Affiliations
Contributions
LY and WJ contributed to the conception and design of the study, LY, LX, GC, MH and ZX contributed to sample recruitment and patient clinical evaluation, FF, LX, GC MH and ZX conducted the experiment, WJ, FF and MH organized the database and performed the statistical analysis, FF wrote the first draft of the manuscript, and WJ wrote sections of the manuscript. All authors contributed to manuscript revisions and read and approved the submitted version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the Eye and ENT Hospital of Fudan University. All research was conducted in accordance with the Declaration of Helsinki. All participants (parents on behalf of their children) provided written informed consent forms for both genetic counselling and molecular genetic testing prior to enrolment.
Consent for publication
All participants (parents on behalf of their children) of the study gave written consent for their personal or clinical details along with any identifying images to be published in this study.
Competing interests
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1:
Table S1. Gene list of capture panel. 792 genes most frequently involved in common inherited eye diseases.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Fan, F., Luo, Y., Wu, J. et al. The mutation spectrum in familial versus sporadic congenital cataract based on next-generation sequencing. BMC Ophthalmol 20, 361 (2020). https://doi.org/10.1186/s12886-020-01567-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12886-020-01567-x