Abstract
Background
To compare the diagnostic performance of neurite orientation dispersion and density imaging (NODDI), mean apparent propagator magnetic resonance imaging (MAP-MRI), diffusion kurtosis imaging (DKI), diffusion tensor imaging (DTI) and diffusion-weighted imaging (DWI) in distinguishing high-grade gliomas (HGGs) from solitary brain metastases (SBMs).
Methods
Patients with previously untreated, histopathologically confirmed HGGs (n = 20) or SBMs (n = 21) appearing as a solitary and contrast-enhancing lesion on structural MRI were prospectively recruited to undergo diffusion-weighted MRI. DWI data were obtained using a q-space Cartesian grid sampling procedure and were processed to generate parametric maps by fitting the NODDI, MAP-MRI, DKI, DTI and DWI models. The diffusion metrics of the contrast-enhancing tumor and peritumoral edema were measured. Differences in the diffusion metrics were compared between HGGs and SBMs, followed by receiver operating characteristic (ROC) analysis and the Hanley and McNeill test to determine their diagnostic performances.
Results
NODDI-based isotropic volume fraction (Viso) and orientation dispersion index (ODI); MAP-MRI-based mean-squared displacement (MSD) and q-space inverse variance (QIV); DKI-generated radial, mean diffusivity and fractional anisotropy (RDk, MDk and FAk); and DTI-generated radial, mean diffusivity and fractional anisotropy (RD, MD and FA) of the contrast-enhancing tumor were significantly different between HGGs and SBMs (p < 0.05). The best single discriminative parameters of each model were Viso, MSD, RDk and RD for NODDI, MAP-MRI, DKI and DTI, respectively. The AUC of Viso (0.871) was significantly higher than that of MSD (0.736), RDk (0.760) and RD (0.733) (p < 0.05).
Conclusion
NODDI outperforms MAP-MRI, DKI, DTI and DWI in differentiating between HGGs and SBMs. NODDI-based Viso has the highest performance.
Similar content being viewed by others
Background
High-grade gliomas (HGGs) and brain metastases are common malignancies in the central nervous system (CNS). HGGs account for approximately 80% of primary CNS malignancies [1]. Meanwhile, metastatic tumors occur ten times more frequently than primary malignancy in the brain [2]. The differentiation between HGGs and brain metastases is critical, as the management strategies for these two malignant brain tumors are vastly different. For patients with HGGs, surgical resection is the first choice, and it is usually not necessary to perform a systemic examination [2]. However, for patients with suspected brain metastases, comprehensive systemic examinations are needed, and if confirmed, stereotactic radiosurgery or systemic therapy such as targeted therapy and immunotherapy are recommended [3].
Magnetic resonance imaging (MRI) is the mainstay of imaging modalities for the diagnosis of brain tumors. For patients who present multiple cerebral lesions and have a history of primary malignancy, the diagnosis of brain metastases may be straightforward by MRI. However, solitary brain metastases (SBMs) are the first manifestation in nearly 30% of patients with systemic malignancy [4]. Therefore, when patients show a solitary and contrast-enhancing brain lesion, it would be challenging to distinguish HGG from solitary brain metastasis (SBM) because they often show similar signal features and contrast enhancement patterns on conventional MRI, leading to incorrect diagnosis in over 40% of cases [5]. In this case, tumor biopsy is often performed to confirm the histologic diagnosis, whereas it has inherent limitations, such as procedure-related complications, interobserver variability and sampling errors [6]. Thus, a noninvasive method to differentiate HGGs from SBMs is preferable and sometimes mandatory when the patient cannot receive surgery due to poor general condition or when the tumor involves or is adjacent to important brain areas.
Diffusion-weighted imaging (DWI) is one of the most widely used advanced MRI techniques to characterize the microstructural changes in cerebral tumors, which complements the anatomic information provided by conventional MRI [7]. Previously, Gaussian-based DWI and diffusion tensor imaging (DTI) have been used to distinguish HGGs from SBMs, with DWI-based apparent diffusion coefficient (ADC) and DTI-generated fractional anisotropy (FA) being the most commonly used metrics. However, contradictory results have been reported on the ability of ADC and FA to differentiate HGGs from SBMs [8,9,10].
Recently, novel diffusion MRI techniques, such as the three-compartment biophysical model neurite orientation dispersion and density imaging (NODDI) and the non-Gaussian-based mean apparent propagator (MAP)-MRI, have emerged as powerful tools to evaluate brain microstructure in vivo, as they can provide new insights into the complexity and inhomogeneity of brain microstructure [11, 12]. Both NODDI and MAP-MRI have shown promising results in lateralization of temporal lobe epilepsy [13], assessment of Parkinson’s disease [14] and grading of gliomas [15]; nonetheless, whether they outperformed the more commonly used non-Gaussian-based DKI and Gaussian diffusion models such as DTI and DWI in differentiation between HGGs and SBMs remains unknown. Therefore, the aim of our study was to compare the diagnostic performance of NODDI, MAP-MRI, DKI, DTI and DWI in distinguishing HGGs from SBMs.
Methods
Study participants
Our institutional review board approved this prospective study, and all participants provided written informed consent. From January 2019 to March 2020, 175 consecutive patients who presented a solitary and contrast-enhancing brain lesion on structural MRI, which were identified by a radiologist with 3 years of experience, were enrolled to undergo diffusion-weighted MRI. The inclusion criteria were as follows: (a) a solitary and contrast-enhancing brain lesion on structural MRI, and (b) a pathological diagnosis of HGG according to the world health organization (WHO) 2016 classification of brain tumors, or a pathological diagnosis of SBM. The exclusion criteria were as follows: (a) brain lesions that had received previous treatment before MRI, (b) brain lesions that had received no surgery or biopsy after MRI, (c) histologically confirmed other diseases except HGG and SBM, and (d) poor-quality MR images due to movement artifacts. Finally, 41 patients (26 males and 15 females; mean age, 54.85 years; age, 19–81 years) were included in our study. The flowchart for the selection of the study population is shown in Fig. 1.
MRI
All patients included in our study underwent structural and diffusion MRI on a 3.0T scanner (MAGNETOM Skyra, Siemens Healthcare, Erlangen, Germany) with a 20-channel head/neck coil. The structural MRI sequences included axial turbo spin echo (TSE) T2-weighted (T2W) imaging and axial TSE T1-weighted (T1W) imaging. After intravenous administration of 0.1 mmol/kg gadobutrol (Gadovist, Bayer Healthcare), axial contrast-enhanced TSE T1W imaging, as well as coronal and sagittal contrast-enhanced FLASH T1W imaging, was performed. On the second day after the structural MRI, diffusion MRI was performed in the axial plane using a full q-space Cartesian grid sampling procedure with a radial grid size of 3, ninety-nine diffusion directions and ten different b-values (from 0 to 3000 s/mm2). The acquisition time was 10 min 32 s. The acquisition parameters of all MR sequences are shown in Table 1. The geometric parameters, including slice thickness, slice gap and FOV of axial T1W and T2W, were identical to those of diffusion-weighted MRI. Moreover, the imaging planes of axial T2W and T1W and diffusion MRI were all aligned parallel to the genu and splenium line of the corpus callosum.
Diffusion data analysis
All DWI data were converted to the NIfTI format using the DCM2NII tool and then processed using the NeuDiLab software developed in-house based on the open-resource tool DIPY (Diffusion Imaging in Python, http://nipy.org/dipy). The five diffusion models and the derived diffusion metrics are as follows:
For the conventional DWI model, the ADC measures the magnitude of diffusion of water molecules in a voxel, which was calculated with the following equation [16]:
where S(b) is the signal intensity according to the given b value and S (0) is the signal intensity for b = 0 s/mm2.
For the DTI model, the axial and radial diffusivity (AD and RD) are the average diffusivities, respectively, in the directions parallel and perpendicular to the diffusion tensor (DT) eigenvector with the largest eigenvalue. Mean diffusivity (MD) quantifies the mean extent of the diffusion of water molecules in a voxel and reflects the overall level of molecular dispersion. Fractional anisotropy (FA) represents the amount of diffusion asymmetry within a voxel, which in theory should range between 0 and 1. AD, RD, MD and FA were all derived from the principal eigenvalues (λ1, λ2, and λ3) of DT with the following equations [17]:
For the DKI model, AD, RD, MD, and FA were denoted as ADk, RDk, MDk, and FAk, respectively, which were derived from the principal eigenvalues (λ1, λ2, and λ3) of the corrected diffusion tensor (DT) with the same equations as the DTI model. The axial, radial, and mean kurtosis (AK, RK, and MK) were derived from the kurtosis tensor (KT). Specifically, AK and RK are the average kurtosis parallel and perpendicular to the principle diffusion eigenvector. MK is the average kurtosis of all diffusion directions. The DKI model is described with the following equation [18]:
where ADCDKI is the apparent diffusion coefficient obtained with DKI and K is the kurtosis parameter.
For the MAP model, the non-Gaussianity (NG) characterizes the three-dimensional diffusion process and is defined as \(NG = \sin \uptheta {\text{PG}}\), which quantifies the dissimilarity between the propagator, P(r), and its Gaussian part, G(r). Axial non-Gaussianity (NGAx) and radial non-Gaussianity (NGRad) are the derivations of NG for diffusion on the axial and radial directions, respectively. The return-to-origin probability (RTOP) describes the probability of no net displacement of molecules between two diffusion sensitization gradients, and the return-to-plane probability (RTPP) and return-to-axis probability (RTAP) are its variants for diffusion in one- and two-dimensions. The mean squared displacement (MSD) measures the average amount of diffusion in a voxel. The q-space inverse variance (QIV) measures the inverse variance of the q-signal geometric means. RTOP, RTAP, RTPP, MSD and QIV are calculated using the following equations [19]:
where q indicates the q-space wave-vector; E(q) is the ratio of the signal at q to that at q = 0; q⊥ denotes the q-vector on the sampled plane; and q// denotes the component of the q-vector along the fiber axis. R denotes a three-dimensional displacement vector. P(R) indicates the likelihood of particles to undergo a net displacement.
For the NODDI model, the isotropic volume fraction (Viso) measures the isotropic diffusion compartment in a voxel, the intracellular volume fraction (Vic) represents diffusion within the axons and cells, and the orientation dispersion index (ODI) measures the orientation dispersion of fibers in a voxel. Viso, Vic and ODI are calculated using the following equation [17]:
where Eic is the signal contribution from the intracellular compartment; Eec is the signal due to diffusion in the extracellular space; and an isotropic Gaussian compartment Eiso represents free diffusion.
All diffusion parametric images and axial postcontrast T1W images were coregistered to T2W images using Elastix software (http://elastix.isi.uu.nl/) and transformed into the same imaging space. For diffusion images, baseline data with b = 0 were used for registration. Rigid transform was used due to its robustness with comparison to affine transform for the multicontrast data. For quantitative analysis, a neuroradiologist (with 8 years of experience in neuroradiology) performed region of interest (ROI) measurements with guidance from a board-certified radiologist (with 17 years of experience in neuroradiology), and both radiologists were blinded to the histological results. All the ROIs were drawn on the registered postcontrast T1W images and T2W images using the open-source application ITK-SNAP (www.itk-snap.org). The enhanced areas seen on the postcontrast T1W images were delineated and defined as the ROIs of the contrast-enhancing tumor. The hyperintense signal that represented peritumoral edema on the T2W images was manually outlined and defined as the ROI of peritumoral edema. Areas of necrosis, cysts or hemorrhage that were detectable on the postcontrast T1W images or T2W images were excluded from the ROIs. Finally, the ROIs were directly copied to the coregistered parametric diffusion maps of the same patient by using MRIcron software (https://people.cas.sc.edu/rorden/mricron/index.html) to calculate the corresponding average values of the contrast-enhancing tumor and peritumoral edema.
Statistical analysis
The normality and equal variance of diffusion metrics were checked using the Shapiro–Wilk test and Levene’s F test, respectively. Differences in all diffusion parameters between HGGs and SBMs were compared by using the independent Student’s t test or the Mann–Whitney U test. The diagnostic performance of the significant diffusion metrics to discriminate HGGs from SBMs was evaluated by receiver operating characteristic (ROC) curve analysis. The optimal cutoff value, sensitivity, specificity and accuracy were calculated. The area under the curve (AUC) was compared by using the Hanley and McNeill test using the R software (version 3.2.4; R Foundation for Statistical Computing). All other statistical analyses were performed using SPSS (version 26.0; SPSS, Chicago, III, USA). A two-tailed p < 0.05 was considered to indicate a significant difference.
Results
Study population
Twenty patients (13 males and 7 females; mean age, 55.70 years; age, 19–67 years) with HGGs diagnosed by histopathology including seven patients with anaplastic astrocytoma (WHO grade III) and thirteen patients with glioblastoma (WHO grade IV) and twenty-one patients (13 males and 8 females; mean age, 54.05 years; age, 43–81 years) with SBMs confirmed by histopathology were included. The primary tumors were lung carcinoma (n = 10), breast carcinoma (n = 5), colon carcinoma (n = 3), liver carcinoma (n = 1), gastric carcinoma (n = 1), and thyroid carcinoma (n = 1).
Diffusion parameters between HGGs and SBMs
The averages of all diffusion parameters and their comparison between the HGG group and SBM group are shown in Table 2. The RD, MD, RDk, MDk, MSD, QIV, Viso and ODI of the contrast-enhancing tumors were significantly lower in the HGGs than in the SBMs (p = 0.006, p = 0.016, p = 0.002, p = 0.007, p = 0.002, p = 0.039, p = 0.001, and p = 0.003, respectively). The FA and FAk of the contrast-enhancing tumors were significantly higher in the HGGs than in the SBMs (p = 0.007 and p = 0.021, respectively). No significant differences were found among all other diffusion parameters in the contrast-enhancing tumors or peritumoral edema between the two groups (p > 0.05).
Diagnostic performances of diffusion metrics
ROC curve analyses of the significant diffusion metrics of the contrast-enhancing tumors are shown in Table 3 and Fig. 2. The best single discriminative parameters for DTI, DKI, MAP-MRI and NODDI were RD, RDk, MSD and Viso, respectively (AUC = 0.733, 0.760, 0.736, and 0.871, respectively). Among them, NODDI-based Viso showed the best performance in differentiating HGGs from SBMs, with a sensitivity of 95.0%, specificity of 76.2% and accuracy of 85.4% at the optimal threshold of 0.158. The AUC of NODDI-based Viso was significantly higher than MAP-MRI-based MSD, DKI-based RDk and DTI-based RD (p = 0.012; p = 0.047; p = 0.007), suggesting that NODDI outperforms MAP-MRI, DKI and DTI in distinguishing HGGs from SBMs. No significant differences were found between the AUCs of RD, RDk and MSD (p > 0.05). Two representative cases in each group are shown in Figs. 3 and 4.
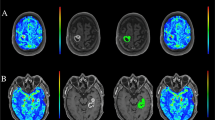
A patient with histologically confirmed glioblastoma. a T2W images and b post-contrast T1W images show a contrast-enhancing tumor with peritumoral edema located in the right frontal lobe. Pseudocolorful maps show the lesion (inside the white circle) having a slightly increased RD (c), RDk (d), MSD (e), and Viso (f) compared to the contralateral normal white matter
A patient with a histologically confirmed solitary brain metastasis from the colon carcinoma. a T2W images and b post-contrast T1W images represent a contrast-enhancing tumor with peritumoral edema located in the right parietal lobe. Pseudocolorful maps show the lesion (inside the white circle) having a slightly increased RD (c), RDk (d), MSD (e) and a moderately increased Viso (f) compared to the contralateral normal white matter
Discussion
Our results demonstrated that HGGs and SBMs showed distinctive NODDI, MAP-MRI, DKI and DTI-based diffusion metrics in the contrast-enhancing tumor region, while no difference was observed for any of the diffusion parameters in peritumoral edema. NODDI-based tumoral Viso had the greatest discriminative power between HGGs and SBMs.
Patients with HGGs or SBMs generally have a dismal prognosis, but correct differential diagnosis and appropriate clinical decisions can significantly prolong the survival time [20]. Thus, it is crucial to distinguish HGGs from SBMs. Nonetheless, it is always challenging to differentiate between these two malignancies only by conventional MRI [5]. In recent years, advanced diffusion-weighted MRI techniques have emerged as powerful tools to assess microstructural changes in CNS diseases. As the complex microstructures in neural tissue (e.g., cell membranes and myelin fibers) change water molecule diffusion into a non-Gaussian probability distribution, non-Gaussian diffusion models such as MAP-MRI and DKI are supposed to reflect the real situation of water molecule diffusion more accurately and better characterize the complexity and inhomogeneity of the tissue microenvironment than Gaussian diffusion models [21]. Specifically, DKI is a commonly used and moderately complex physical model that is sensitive to DWI sampling and noise [22]. MAP-MRI is a more recent and highly complex physical model that can evaluate three-dimensional q-space data [11] but shows increased sensitivity to DWI sampling and noise [22]. Comparatively, NODDI is an increasingly popular biophysical model with low complexity that attempts to separate the signal contribution of neural tissue into three compartments, including restricted, hindered, and isotropic diffusion, and to model the dispersion of axonal fibers [17]. NODDI metrics are highly stable to DWI sampling and image quality [22]. In the present study, we found that NODDI-based Viso outperformed other non-Gaussian or Gaussian diffusion parameters in the differentiation between HGGs and SBMs. Although no single diffusion parameter can fully capture the complexity of neural tissue, our findings suggest that NODDI-Viso could potentially be a sensitive imaging biomarker in neuro-oncology research and deserves further investigation.
NODDI-Viso represents isotropic diffusion within the tissue; in our study, HGGs showed a lower tumoral Viso value than SBMs. This phenomenon can be explained by the fact that HGGs are characterized by enlarged extracellular space and overproduction of certain components of extracellular matrix components, mainly tenascin [23]. These molecules accumulate and orient in the extracellular matrix [24], resulting in less isotropy at DWI. In contrast, metastatic brain tumors degrade the extracellular matrix with heparanase and matrix metalloproteinases, thereby growing into the brain parenchyma in an expansive and noninfiltrating pattern [25], resulting in higher isotropy at DWI. NODDI-based ODI represents the orientation dispersion of fibers in tissue [12]. In this study, the tumoral ODI value was found to be lower in HGGs than in SBMs, which could also be explained by the fact that tumor tissue tends to be less isotropic for HGGs than for SBMs. For MAP-MRI, we found that HGGs showed lower MSD and QIV values than SBMs. MSD indicates the mean square displacement of the water molecules. These results can be explained by the fact that the solid part of HGGs had higher cellularity than did brain metastases [26,27,28], which might lead to higher diffusion restriction in HGGs, and the water molecules will thus move shorter distances and result in a lower MSD value [19]. QIV signifies the q-space inverse variance, which is a pseudodiffusivity measure and represents different diffusion components [29]; thus, a higher tumoral QIV value for SBM suggested a higher proportion of fast diffusivity in SBMs. For DTI, we found that HGGs showed a higher tumoral FA value and lower tumoral RD and MD values than SBMs. FA is a measurement of the directionality of water diffusion along with the white matter, which has a positive correlation with tumor cellularity [30]. Higher tumoral FA values for HGGs than SBMs were also described in recent studies [31, 32], where a higher FA value of the contrast-enhancing region of HGGs was reported to be assumed to be due to the higher cellularity of HGGs [26,27,28]. MD reveals the rate of water molecule diffusional motion; RD represents the diffusion rate of water perpendicular to white matter fibers [17]. Both MD and RD show an inverse relationship with tumor cellularity [33, 34], which can explain the opposite change trend of MD and RD compared with FA. As an extension of DTI [35], DKI-based RD, MD and FA in our study showed similar change patterns with DTI-based RD, MD and FA.
Previously, the ability of diffusion MR metrics such as ADC and FA to distinguish peritumoral edema of HGGs from that of SBMs has been widely investigated, but the study results remain controversial [8,9,10]. In the present study, although five diffusion models were utilized, no significant differences were found in any of the diffusion parameters between the peritumoral edema of HGGs and SBMs. These inconsistent results may contribute to the intrinsic heterogeneity of HGGs. Although it was confirmed histologically that tumor cells exist in the peritumoral edema of HGGs, the magnitude of tumor cell infiltration actually has a substantially wide range [36]. Thus, minimal tumor infiltration may not cause a significant signal change in diffusion MRI. Further studies applying three-dimensional texture analysis of volumetric diffusion MR images could provide additional information on the heterogeneity of tumor cell infiltration in peritumoral edema, which may be helpful in the differentiation of tumor-infiltrated edema from purely vasogenic edema.
Our study has some limitations. First, the sample size was small for both the HGG and SBM groups, as all patients were prospectively enrolled from a single institution. However, our study showed that the advanced diffusion-weighted technique NODDI-based Viso had a desirable diagnostic performance (AUC = 0.871) for distinguishing HGGs from SBMs. This model deserves further study with a larger sample size to validate the current results. Second, the diffusion MR examination in our study requires a long scan duration of approximately 10 min. In the future, this problem can be overcome by using advanced techniques, such as compressed sensing [37] and simultaneous multislice acquisition techniques [38].
Conclusion
Our study shows that NODDI outperforms MAP-MRI, DKI, DTI and DWI in distinguishing HGGs from SBMs. Among all the diffusion metrics, NODDI-based Viso has the highest performance in differentiating between HGGs and SBMs.
Availability of data and materials
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- NODDI:
-
Neurite orientation dispersion and density
- MAP:
-
Mean apparent propagator
- MRI:
-
Magnetic resonance imaging
- DKI:
-
Diffusion kurtosis imaging
- DTI:
-
Diffusion tensor imaging
- DWI:
-
Diffusion-weighted imaging
- HGG:
-
High-grade glioma
- SBM:
-
Solitary brain metastasis
- ROC:
-
Receiver operating characteristic
- Viso :
-
Isotropic volume fraction
- ODI:
-
Orientation dispersion index
- MSD:
-
Mean-squared displacement
- QIV:
-
Q-space inverse variance
- RDk :
-
DKI-generated radial diffusivity
- MDk :
-
DKI-generated mean diffusivity
- FAk :
-
DKI-generated fractional anisotropy
- RD:
-
Radial diffusivity
- MD:
-
Mean diffusivity
- FA:
-
Fractional anisotropy
- CNS:
-
Central nervous system
- ADC:
-
Apparent diffusion coefficient
- WHO:
-
World Health Organization
- TSE:
-
Turbo spin echo
- T2W:
-
T2-weighted
- T1W:
-
T1-weighted
- AD:
-
Axial diffusivity
- DT:
-
Diffusion tensor
- ADk :
-
DKI-generated axial diffusivity
- AK:
-
Axial kurtosis
- RK:
-
Radial kurtosis
- MK:
-
Mean kurtosis
- KT:
-
Kurtosis tensor
- NG:
-
Non-Gaussianity
- NGAx :
-
Axial non-Gaussianity
- NGRad :
-
Radial non-Gaussianity
- RTOP:
-
Return-to-origin probability
- RTPP:
-
Return-to-plane probability
- RTAP:
-
Return-to-axis probability
- Vic :
-
Intracellular volume fraction
- ROI:
-
Region of interest
- AUC:
-
Area under the curve
References
Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro-oncology. 2018;20(suppl_4):iv1–86.
Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet (London, England). 2018;392(10145):432–46.
Lin X, DeAngelis LM. Treatment of brain metastases. J Clin Oncol. 2015;33(30):3475–84.
Schiff D. Single brain metastasis. Curr Treat Options Neurol. 2001;3(1):89–99.
Blanchet L, Krooshof PW, Postma GJ, Idema AJ, Goraj B, Heerschap A, Buydens LM. Discrimination between metastasis and glioblastoma multiforme based on morphometric analysis of MR images. AJNR Am J Neuroradiol. 2011;32(1):67–73.
Jackson RJ, Fuller GN, Abi-Said D, Lang FF, Gokaslan ZL, Shi WM, Wildrick DM, Sawaya R. Limitations of stereotactic biopsy in the initial management of gliomas. Neuro-oncology. 2001;3(3):193–200.
Svolos P, Kousi E, Kapsalaki E, Theodorou K, Fezoulidis I, Kappas C, Tsougos I. The role of diffusion and perfusion weighted imaging in the differential diagnosis of cerebral tumors: a review and future perspectives. Cancer Imaging. 2014;14(1):20.
Caravan I, Ciortea CA, Contis A, Lebovici A. Diagnostic value of apparent diffusion coefficient in differentiating between high-grade gliomas and brain metastases. Acta Radiol. 2018;59(5):599–605.
van Westen D, Lätt J, Englund E, Brockstedt S, Larsson EM. Tumor extension in high-grade gliomas assessed with diffusion magnetic resonance imaging: values and lesion-to-brain ratios of apparent diffusion coefficient and fractional anisotropy. Acta Radiol. 2006;47(3):311–9.
Han C, Huang S, Guo J, Zhuang X, Han H. Use of a high b-value for diffusion weighted imaging of peritumoral regions to differentiate high-grade gliomas and solitary metastases. J Magn Reson Imaging. 2015;42(1):80–6.
Özarslan E, Koay CG, Shepherd TM, Komlosh ME, İrfanoğlu MO, Pierpaoli C, Basser PJ. Mean apparent propagator (MAP) MRI: a novel diffusion imaging method for mapping tissue microstructure. Neuroimage. 2013;78:16–32.
Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage. 2012;61(4):1000–16.
Ma K, Zhang X, Zhang H, Yan X, Gao A, Song C, Wang S, Lian Y, Cheng J. Mean apparent propagator-MRI: a new diffusion model which improves temporal lobe epilepsy lateralization. Eur J Radiol. 2020;126:108914.
Mitchell T, Archer DB, Chu WT, Coombes SA, Lai S, Wilkes BJ, McFarland NR, Okun MS, Black ML, Herschel E, et al. Neurite orientation dispersion and density imaging (NODDI) and free-water imaging in Parkinsonism. Hum Brain Mapp. 2019;40(17):5094–107.
Zhao J, Li JB, Wang JY, Wang YL, Liu DW, Li XB, Song YK, Tian YS, Yan X, Li ZH, et al. Quantitative analysis of neurite orientation dispersion and density imaging in grading gliomas and detecting IDH-1 gene mutation status. Neuroimage Clin. 2018;19:174–81.
Le Bihan D, Breton E, Lallemand D, Aubin ML, Vignaud J, Laval-Jeantet M. Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging. Radiology. 1988;168(2):497–505.
Kodiweera C, Alexander AL, Harezlak J, McAllister TW, Wu YC. Age effects and sex differences in human brain white matter of young to middle-aged adults: a DTI, NODDI, and q-space study. Neuroimage. 2016;128:180–92.
Tabesh A, Jensen JH, Ardekani BA, Helpern JA. Estimation of tensors and tensor-derived measures in diffusional kurtosis imaging. Magn Reson Med. 2011;65(3):823–36.
Fick RHJ, Wassermann D, Caruyer E, Deriche R. MAPL: tissue microstructure estimation using Laplacian-regularized MAP-MRI and its application to HCP data. Neuroimage. 2016;134:365–85.
Kaal EC, Niël CG, Vecht CJ. Therapeutic management of brain metastasis. Lancet Neurol. 2005;4(5):289–98.
Jensen JH, Helpern JA. MRI quantification of non-Gaussian water diffusion by kurtosis analysis. NMR Biomed. 2010;23(7):698–710.
Hutchinson EB, Avram AV, Irfanoglu MO, Koay CG, Barnett AS, Komlosh ME, Özarslan E, Schwerin SC, Juliano SL, Pierpaoli C. Analysis of the effects of noise, DWI sampling, and value of assumed parameters in diffusion MRI models. Magn Reson Med. 2017;78(5):1767–80.
Zamecnik J. The extracellular space and matrix of gliomas. Acta Neuropathol. 2005;110(5):435–42.
Pope WB, Mirsadraei L, Lai A, Eskin A, Qiao J, Kim HJ, Ellingson B, Nghiemphu PL, Kharbanda S, Soriano RH, et al. Differential gene expression in glioblastoma defined by ADC histogram analysis: relationship to extracellular matrix molecules and survival. AJNR Am J Neuroradiol. 2012;33(6):1059–64.
Preusser M, Capper D, Ilhan-Mutlu A, Berghoff AS, Birner P, Bartsch R, Marosi C, Zielinski C, Mehta MP, Winkler F, et al. Brain metastases: pathobiology and emerging targeted therapies. Acta Neuropathol. 2012;123(2):205–22.
Altman DA, Atkinson DS Jr, Brat DJ. Best cases from the AFIP: glioblastoma multiforme. Radiographics. 2007;27(3):883–8.
Tsougos I, Svolos P, Kousi E, Fountas K, Theodorou K, Fezoulidis I, Kapsalaki E. Differentiation of glioblastoma multiforme from metastatic brain tumor using proton magnetic resonance spectroscopy, diffusion and perfusion metrics at 3 T. Cancer Imaging. 2012;12(3):423–36.
Carrier DA, Mawad ME, Kirkpatrick JB, Schmid MF. Metastatic adenocarcinoma to the brain: MR with pathologic correlation. AJNR Am J Neuroradiol. 1994;15(1):155–9.
Hosseinbor AP, Chung MK, Wu YC, Alexander AL. Bessel Fourier orientation reconstruction (BFOR): an analytical diffusion propagator reconstruction for hybrid diffusion imaging and computation of q-space indices. Neuroimage. 2013;64:650–70.
Kinoshita M, Hashimoto N, Goto T, Kagawa N, Kishima H, Izumoto S, Tanaka H, Fujita N, Yoshimine T. Fractional anisotropy and tumor cell density of the tumor core show positive correlation in diffusion tensor magnetic resonance imaging of malignant brain tumors. Neuroimage. 2008;43(1):29–35.
Bette S, Huber T, Wiestler B, Boeckh-Behrens T, Gempt J, Ringel F, Meyer B, Zimmer C, Kirschke JS. Analysis of fractional anisotropy facilitates differentiation of glioblastoma and brain metastases in a clinical setting. Eur J Radiol. 2016;85(12):2182–7.
Wang S, Kim SJ, Poptani H, Woo JH, Mohan S, Jin R, Voluck MR, O’Rourke DM, Wolf RL, Melhem ER, et al. Diagnostic utility of diffusion tensor imaging in differentiating glioblastomas from brain metastases. AJNR Am J Neuroradiol. 2014;35(5):928–34.
Yuan W, Holland SK, Jones BV, Crone K, Mangano FT. Characterization of abnormal diffusion properties of supratentorial brain tumors: a preliminary diffusion tensor imaging study. J Neurosurg Pediatr. 2008;1(4):263–9.
Marini C, Iacconi C, Giannelli M, Cilotti A, Moretti M, Bartolozzi C. Quantitative diffusion-weighted MR imaging in the differential diagnosis of breast lesion. Eur Radiol. 2007;17(10):2646–55.
Hui ES, Cheung MM, Qi L, Wu EX. Towards better MR characterization of neural tissues using directional diffusion kurtosis analysis. Neuroimage. 2008;42(1):122–34.
Yamahara T, Numa Y, Oishi T, Kawaguchi T, Seno T, Asai A, Kawamoto K. Morphological and flow cytometric analysis of cell infiltration in glioblastoma: a comparison of autopsy brain and neuroimaging. Brain Tumor Pathol. 2010;27(2):81–7.
Arai K, Belthangady C, Zhang H, Bar-Gill N, DeVience SJ, Cappellaro P, Yacoby A, Walsworth RL. Fourier magnetic imaging with nanoscale resolution and compressed sensing speed-up using electronic spins in diamond. Nat Nanotechnol. 2015;10(10):859–64.
Lee HL, Li Z, Coulson EJ, Chuang KH. Ultrafast fMRI of the rodent brain using simultaneous multi-slice EPI. Neuroimage. 2019;195:48–58.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. U1801681; 82001768), Key Areas Research and Development Program of Guangdong (Grant No. 2019B020235001), Fundamental Research Funds for the Central Universities (Grant No. 20ykpy102), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2017), Guangdong Basic and Applied Basic Research Foundation (Grant No. 2019A1515110189), Guangdong Natural Science Foundation (Grant No. 2017A030313777), Medical science and Technology Research Fund of Guangdong Province (Grant No. A2020074). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
JM: Conceptualization, Methodology. WZ: Resources, Investigation. QZ and ZY: Investigation, Formal analysis. XY, HZ, MW, GY and MZ: Software, Data curation, Formal analysis. JS: Project administration, Supervision. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This prospective study was approved by the institutional research ethics board of the Institutional Review Board of Sun Yat-Sen Memorial Hospital of Sun Yat-Sen University, and written informed consent was obtained from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Mao, J., Zeng, W., Zhang, Q. et al. Differentiation between high-grade gliomas and solitary brain metastases: a comparison of five diffusion-weighted MRI models. BMC Med Imaging 20, 124 (2020). https://doi.org/10.1186/s12880-020-00524-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12880-020-00524-w