Abstract
Background
The increasing prevalence of methicillin-resistant Staphylococcus aureus (MRSA) strains resistant to non-beta-lactam antimicrobials poses a significant challenge in treating severe MRSA bloodstream infections. This study explores resistance development and mechanisms in MRSA isolates, especially after the first dalbavancin-resistant MRSA strain in our hospital in 2016.
Methods
This study investigated 55 MRSA bloodstream isolates (02/2015–02/2021) from the University Hospital of the Medical University of Vienna, Austria. The MICs of dalbavancin, linezolid, and daptomycin were assessed. Two isolates (16–33 and 19–362) resistant to dalbavancin were analyzed via whole-genome sequencing, with morphology evaluated using transmission electron microscopy (TEM).
Results
S.aureus BSI strain 19–362 had two novel missense mutations (p.I515M and p.A606D) in the pbp2 gene. Isolate 16–33 had a 534 bp deletion in the DHH domain of GdpP and a SNV in pbp2 (p.G146R). Both strains had mutations in the rpoB gene, but at different positions. TEM revealed significantly thicker cell walls in 16–33 (p < 0.05) compared to 19–362 and dalbavancin-susceptible strains. None of the MRSA isolates showed resistance to linezolid or daptomycin.
Conclusion
In light of increasing vancomycin resistance reports, continuous surveillance is essential to comprehend the molecular mechanisms of resistance in alternative MRSA treatment options. In this work, two novel missense mutations (p.I515M and p.A606D) in the pbp2 gene were newly identified as possible causes of dalbavancin resistance.
Similar content being viewed by others
Background
Methicillin-resistant Staphylococcus aureus (MRSA) is a Gram positive, coagulase-positive bacterium of the Staphylococcaceae family, which is resistant to beta-lactam antimicrobials due to the penicillin-binding protein 2a (PBP2a). Common diseases caused by MRSA include skin and soft tissue infections, bone and joint infections, pneumonia as well as bloodstream infections (BSI) [1]. Bacteremia leads to a high morbidity and fatality rate of 20–25% [2]. Vancomycin has been the first line therapy in MRSA bacteremia and endocarditis for the last decades [3]. According to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) 2023, all Staphylococcus (S.) aureus isolates with minimum inhibitory concentrations (MICs) > 2 mg/L are considered resistant to vancomycin [4]. Increased use of vancomycin has led to the development of MRSA with reduced susceptibility to vancomycin (VISA), due to the adaptability of the pathogen [5]. Vancomycin exerts its bactericidal effect by disrupting bacterial cell wall synthesis. Resistance to vancomycin develops through two main mechanisms: VISA has a thicker, less cross-linked cell wall that traps glycopeptides, and vancomycin-resistant Staphylococcus aureus (VRSA) acquires high-level resistance via the vanA operon from Enterococcus species [6, 7]. Horizontal gene transfer is a major driver for the development of resistance, as well as the subsequent spread. In particular, since horizontal gene transfer gives rise to new and antibiotic-resistant strains, genetic mutation is not a mandatory prerequisite [8].
Because of the increase of vancomycin resistance and the need to improve antimicrobial efficacy and tolerability, other antimicrobials such as the synthetic oxazolidinone linezolid, the cyclic lipopeptide daptomycin, and the semisynthetic lipoglycopeptide dalbavancin, which is derived from the teicoplanin-like A-40,926, were approved [9, 10]. Dalbavancin effects are similar to first generation glycopeptide antimicrobials, but it appears to be highly effective against MRSA. Due to its mechanism of inhibiting peptidoglycan cross-linking, similar to vancomycin, and anchoring in the membrane like daptomycin, making it prone to selecting for cross-resistance [11]. Mutations in WalKR, an essential two-component regulatory system (TCS) regulating autolytic activity and cell division, is linked to vancomycin and daptomycin nonsusceptibility and to dalbavancin resistance in vitro [11, 12]. Since in vivo reports are still very rare, more detailed analyses of resistant strains are necessary. In literature, MRSA resistance to dalbavancin is less than 1% [13,14,15,16]. According to EUCAST 2023, S. aureus isolates with dalbavancin MIC > 0.125 mg and teicoplanin MIC > 2 mg/L are considered resistant against the particular antimicrobial [4].
In order to find a suitable antimicrobial and to ensure a successful therapy, the Division of Clinical Microbiology at the Medical University of Vienna routinely carries out antimicrobial susceptibility testing (AST) by disk diffusion according to current EUCAST regulations. However, dalbavancin and daptomycin are not part of routine AST, which results in a lack of information about their development of MIC values and resistance. In this study, we examined two dalbavancin-resistant MRSA strains in more detail by performing whole genome sequencing (WGS) and transmission electron microscopy (TEM) to evaluate possible genetic causes of resistance and their morphological impact.
Methods
Sampling
From 01/2015 to 02/2021, a total of 122,560 blood cultures were taken at the Division of Clinical Microbiology at the University Hospital of the Medical University of Vienna (Austria) with 1700 beds. S. aureus was isolated from a total of 1048 blood culture samples, of which 78 (7.4%) were MRSA. Of these, 55 strains were included in this analysis. Only one isolate was collected from each patient, replicates were excluded. Until the start of the analysis, the isolates were stored at -80 °C in a microbank bead-based preservation system (Mast Group Ltd, Merseyside, UK). All isolates were grown on Columbia agar plates supplemented with 5% sheep blood (Becton Dickinson GmbH, Heidelberg, Germany). Matrix-assisted laser desorption/ionization Time-of-flight Mass Spectrometry (MALDI-TOF) (Bruker Daltonics, Bremen, Germany) was performed to verify, that all isolates were S. aureus. The methicillin-resistance of the samples was confirmed by using cefoxitin disk diffusion tests and MRSA ChromAgar (Oxoid Deutschland GmbH, Wesel, Germany). All samples with a cefoxitin inhibition zone diameter < 22 mm were classified as MRSA [4].
Antimicrobial susceptibility testing (AST)
The obtained bacteria suspensions were streaked on Müller-Hinton E Agars (MHE) and MIC gradient test strips (Liofilchem, Roseto degli Abruzzi, Italy) for dalbavancin, linezolid, and daptomycin were added on the agar plates. Before AST with MIC gradient tests strips was performed, the tests were validated using the S. aureus quality control strain ATCC 29,213. Quality controls of the evaluated antimicrobials were within the expected range for all antimicrobials, allowing AST to be performed using MIC gradient test strips. MICs were assessed according to the manufacturer after incubation for 24 h at 35 °C (± 2 °C). The MICs were interpreted according to the EUCAST 2023 guidelines [4]. In addition, a broth microdilution (BMD) assay was performed for resistant isolates in Cation-Adjusted Mueller-Hinton Broth (CA-MHB) plus polysorbate 80 in a final concentration 0.002% [17].
Whole genome sequencing (WGS)
DNA concentration was determined with a Qubit 4.0 (Thermo Fisher Scientific, Waltham, Massachusetts) fluorometer using the dsDNA broad range assay. The Nanodrop 2000c was used to determine the quality/purity of the DNA. The absorbance was measured at 230 nm, 260 nm and 280 nm. A 260/280 absorbance > 1.8 was assumed to be a pure sample free of proteins, and DNA with a 260/230 absorbance ratio > 2.0 was indicated as a pure sample, free of other contaminants such as chloroform or free nucleotides. The preparation of DNA sequencing libraries was conducted using extracted DNA, adhering to the protocol outlined in Illumina DNA Prep®. Following this, the DNA underwent denaturation as specified in the protocol, and was subsequently diluted to reach a final concentration of 8 pM with a 5% PhiX spike-in (PhiX Control v3, Illumina). The sequencing process took place on a V3-flowcell with a 2 × 300 bp configuration using the Illumina MiSeq system (San Diego, CA, USA).
Bioinformatic analysis
For bioinformatic analysis, an inhouse bioinformatics pipeline was used after sequencing to detect mutations. In short, Trim Galore v0.6.5 was used for removing lower quality bases and ensuring a read length of at least 90 bp [18]. Subsequently, reads were mapped to the reference sequence of ATCC 29,213, genbank accession GCA_001879295.1, using Bowtie2 v2.4.2, on top of which VarScan v2.4.4 was used for variant detection [19, 20]. Additionally, a de novo assembly was done with SPAdes v3.15.2 and quality checked using QUAST [21]. For resistance gene detection and genomic characterization of the isolate, the Comprehensive Antibiotic Resistance Database and several tools offered by the Center for Genomic Epidemiology of the National Food Institute of the Technical University of Denmark were used [22, 23].
Transmission electron microscopy
The evaluated isolates were 19–362, 16–33 (both dalbavancin-resistant MRSA strains), ATCC 29,213 (dalbavancin-susceptible control strain), and isolate 15–368 (dalbavancin-susceptible MSSA strain). Isolate 15–368 was collected from the same patient as isolate 16–33 in 2015. TEM analysis was performed as previously described [24]. Briefly, samples were fixed in Karnovsky’s fixative, postfixed in 1% osmium tetroxide and embedded in epoxy resin after dehydration. 70 nm ultra-thin sections were contrasted with 4% neodymium(III)acetate and lead citrate [25]. Images were acquired with Zeiss Libra 120 electron microscope equipped with a bottom mount camera Sharp: eye TRS (2 × 2k) and processed with the ImageSP software. For measurements of the thickness of the cell walls, more than 50 non-dividing cells were imaged for each strain. The ImageSP software package was used for measurements.
Statistical analysis
The determined MICs of dalbavancin, linezolid, and daptomycin were documented in an Excel raw data table, subsequently converted and analyzed with the statistical program “R”. For the comparison of the cell wall thicknesses, to test on normal contribution, a Jarque-Bera test was used. A two-sample t-test assuming equal variances was used for the subsequent analysis.
Results
Antimicrobial susceptibility testing (AST)
AST was performed with with all included 55 MRSA isolates (Table 1). According to EUCAST, two isolates exhibited resistance to dalbavancin [4]. No isolate was resistant to vancomycin.
Minimum inhibitory concentration (MIC) of the evaluated antimicrobials
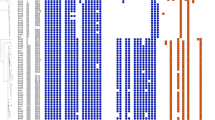
Examination of the dalbavancin, linezolid and daptomycin MICs of all 55 MRSA isolates are shown in Fig. 1. Two isolates with MICs of 0.19 mg/L (19–362) and 0.38 mg/L (16–33) showed resistance to dalbavancin (Fig. 1A) [24]. Further verification of isolate 19–362 was performed by broth microdilution and resulted in an MIC of 0.25 mg/L, in contrast to the MIC strips with an MIC of 0.19 mg/L. Additionally, isolate 19–362 showed resistance to oritavancin (MIC of 0.25 mg/L), was susceptible to teicoplanin (MIC of 1 mg/L) and vancomycin (MIC of 1 mg/L). Isolate 16–33 was susceptible to vancomycin (MIC of 2 mg/L), but resistant to teicoplanin (MIC of 16 mg/L). According to the EUCAST 2023 guidelines, no isolate showed resistance to linezolid or daptomycin [4].
Whole genome sequencing (WGS)
After WGS the MRSA isolate 19–362 with an MIC of 0.19 mg/L, classified as dalbavancin-resistant according to EUCAST 2023 [4], could be assigned to multi locus sequence typing (MLST) ST97, spa type t2297 and SCCmec type V. Additionally one missense mutation (p.N213D) was detected in the ileS gene, which encodes isoleucyl-tRNA synthetase. The isolate had two missense mutations (p.I515M and p.A606D) in the pbp2 gene. Furthermore in isolate 19–362, a missense mutation (p.D530G) was detected in the grlB gene. In addition, a total of seven missense mutations (p.C12F, p.T25A, p.T101R, p.H214C, p.L234H, p.E398A, p.T409A) and one upstream gene variant (n.-54 A > G) were detected in the pbp4 gene. One missense mutation (p.E685K) was found in the rpoB gene. Two missense mutations (p.V72E and p.I97T) occurred in the dfrB gene, which encodes dihydrofolate reductase. Three missense mutations (p.Y410F, p.L450F, and p.V694M) in the grlA gene were detected.
MRSA isolate 16–33, obtained from a blood culture of a patient with cardiac device-related S. aureus endocarditis in 2016, has already been comprehensively analyzed from our working group [24]. Table 2 illustrates the distinct mutations present in isolates 16–33 and 19–362.
Electron microscopic analysis
The dalbavancin-resistant isolates 16–33 and 19–362 exhibited a cell wall thickness of 97.5 nm ± 16.3 (70.0–154.6) and 22.2 nm ± 2.2 (17.7–27.7). In comparison, the dalbavancin-susceptible isolates 15–368 and ATCC 29,213 showed a cell wall thickness of 29.5 nm ± 4.6 (19.2–41.2) and 28.1 nm ± 3.9 (19.7–35.9). The cell wall of 16–33 was significantly (p < 0.001) thicker than the cell wall of all other tested isolates. Interestingly, the second dalbavancin-resistant isolate, 19–362, had the thinnest (p < 0.001) cell wall among all tested isolates. There was no significant difference (p = 0.527) in cell wall thickness between the dalbavancin-susceptible isolate ATCC 29,213 and 15–368 (Fig. 2).
(A) Electron microscopic images of dalbavancin-susceptible (ATCC 29213, 15–368) and dalbavancin-resistant (19–362, 16–33) S. aureus strains. (B) Boxplots of cell wall thickness measurements of dalbavancin-susceptible (ATCC 29213, 15–368) and dalbavancin-resistant (19–362, 16–33) S. aureus strains. The overlayed points indicate individual measurements. Brackets indicate p-values of a Welch t-test, adjusted for multiple testing with Bonferroni correction
Discussion
Dalbavancin
Dalbavancin is a frequently used antimicrobial in the treatment of Gram positive bacterial skin and soft tissue infections, osteomyelitis and endocarditis [1, 2]. Dalbavancin resistance, with a percentage of < 1% of staphylococcal isolates, is considered to be uncommon and without any signs of MIC creep [26]. According to EUCAST 2023, S. aureus isolates with a dalbavancin MIC > 0.125 mg/L are considered resistant [4]. In this analysis, of 55 examined MRSA bloodculture isolates two isolates with MICs of 0.19 mg/L (19–362) and 0.38 mg/L (16–33) were found to be resistant to dalbavancin. The most frequently observed MICs of the 55 MRSA isolates ranged from 0.064 mg/L to 0.094 mg/L. With 147 h to 258 h, dalbavancin has compared to vancomycin a very long half-life [27]. Like vancomycin, it belongs to the lypoglycopeptide antimicrobials. Based on the described reduction in vancomycin susceptibility to MRSA in recent literature, it is valid to assume for dalbavancin to also exhibit reduced susceptibility [28]. Isolate 19–362 showed resistance to oritavancin, another glycopeptide antimicrobial and was susceptible to vancomycin.
As aforementioned, dalbavancin resistance in S. aureus isolates is currently rare and therefore only a few known resistance mechanisms in literature have been reported. These include mutations in ompR, llm, mgtE and a non-synonymous nucleotide substitution in yvgF gene [29], as well as a 534 bp deletion in the DHH domain of GdpP and a single nucleotide variant (SNV) in pbp2 (p. G146R) in isolate 16–33 [24]. In our previous studie, we suggested the SNV in pbp2 as cause for the reported dalbavancin resistant isolate of 16–33 [24].
Dalbavancin unfolds its effect by targeting the bacterial cell wall. It binds to the terminal d-alanyl-d-alanine residues of peptidoglycan precursors, thereby inhibiting catalysis of peptidoglycan crosslinking by the transpeptidase and transglycosylase enzymes. The pbp2 gene is involved in cross-linking of peptidoglycans in the cell wall. Interestingly, isolate 19–362 had two missense mutations in the pbp2 gene. The first (p.I515M) has not been documented yet and the second (p.A606D) has been recently described in a dalbavancin-resistant isolate and was associated with resistance to beta-lactams [30]. In comparison to the 534 bp deletion in the DHH domain of GdpP and a SNV in pbp2 in 16–33, the observed missense mutations in the pbp2 gene in isolate 19–362 did not lead to a thicker cell wall. Alteration of the structural integrity of the cell wall due to mutations within the pbp2 gene could be another cause of dalbavancin resistance, as dalbavancin and other structurally related large molecule antibiotics sterically mediate their anti-infective effect. Hence, a different resistance mechanism might also be imaginable and should be considered in future studies with the identical missense mutations. Furthermore, in light of the previously documented instances of dalbavancin-resistant MRSA strains characterized by the p.A606D missense mutation in the pbp2 gene, it is not possible to assert conclusively that the p.I515N missense mutation exerted a discernible influence on the development of resistance [30]. In addition, mutations in walKR, vraRS, graRS and rpoB have been detected in isolates with reduced glycopeptide susceptibility [31, 32]. Further, walKR-associated gene mutations are associated with dalbavancin resistant strains in vitro [11, 12]. 19–362 did not show mutations in the TCS WalR/WalK regulatory protein, but showed a missense mutation (p.E685K) in the rpoB gene, which encodes the beta subunit of bacterial RNA polymerase, and has been found in previous studies to be a factor in resistance to vancomycin and rifampicin [32,33,34]. Interestingly, despite the missense mutation, 19–362 was not resistant to vancomycin, which is consistent with findings from other studies, in which mutations in the rpoB gene were linked to rifampicin resistance, but did not play a major role in the development of vancomycin-intermediate S. aureus phenotypes [24, 33]. In comparison, 16–33 has two missense mutations in rpoB, but at different positions (p.I560M and p.R483H). Due to the resemblance between dalbavancin and vancomycin, similar resistance mechanisms are conceivable.
Furthermore, in isolate 19–362, a missense mutation (p.D530G) was detected in the grlB gene, which encodes the metabotropic glutamate receptor-like protein B. This missense mutation has not been described before, but there are reports of other mutations in grlB associated with quinolone resistance [35]. The detected missense mutation (p.N213D) in the ileS gene in 19–362, has been reported several times in literature and is associated with mupirocin resistance [36]. The detected missense mutations (p.C12F, p.T25A, p.T101R, p.H214C, p.L234H, p.E398A, p.T409A) and one upstream gene variant (n.-54 A > G) in the pbp4 gene, have already been partially illustrated before [37]. Reduced vancomycin susceptibility is mediated by changes in pbp4 expression [38]. In the dfrB gene, two missense mutations (p.V72E and p.I97T) were detected and are associated with trimethoprim resistance [39]. In the grlA gene, which encodes a DNA topoisomerase and where mutations have previously been linked to quinolone resistance, three missense mutations (p.Y410F, p.L450F, and p.V694M) were detected, of which p.Y410F and p.V964M have already been described [40].
Linezolid and Daptomycin
The linezolid resistance rate for MRSA is currently estimated to be around 0.5%, but a linezolid MIC creep has been reported several times [41]. According to EUCAST 2023, this analysis of 55 isolates, revealed no isolate resistant to linezolid, but ten (18.2%) isolates had an MIC of 2 mg/L [4]. The global prevalence of daptomycin resistance ranges between 0.1 and 0.3%, with an MIC > 1 mg/L [42]. Despite the low prevalence, there are reports, especially in Taiwan, of an MIC creep [43]. In this analysis, none of the examined MRSA isolates was resistant to daptomycin and overall, there was no evidence of an increased MICs between 2015 and 2021.
Limitations
The current study is constrained by the limitation of not having conducted experimental exploration regarding the causal connection between our observed missense mutations in the pbp2 gene and the development of dalbavancin resistance. To address this specific limitation and pave the way for more comprehensive insights, we propose that future studies include experiments involving the transfer of these mutations into an isogenic strain background and measuring the dalbavancin MIC of the mutant and wildtype strain.
Conclusion
Overall, resistance to the non-beta-lactams dalbavancin, daptomycin, and linezolid is still rare in MRSA BSI strains, but due to increasing resistance to vancomycin, the newer antimicrobials should be evaluated intensively, focusing on the underlying molecular mechanisms of resistance. At the University Hospital of the Medical University of Vienna, the three evaluated antimicrobials must be prescribed or approved by an infectious diseases specialist as antibiotic stewardship intervention. A possible reason that only at dalbavancin two resistent strains were found over the years of clinical use could be due to its significantly prolonged half-life compared to daptomycin and linezolid resulting in extended subinhibitory concentrations. This can lead to an increased risk of resistance development.
In this work, two novel missense mutations (p.I515M and p.A606D) in the pbp2 gene that might have led to morphological cell wall alterations in the respective MRSA strain, were newly identified as possible causes of dalbavancin resistance. It should therefore be mentioned that cause of our study’s limitation of not experimentally exploring the causal link between the observed pbp2 gene missense mutations and dalbavancin resistance, future research should involve transferring these mutations into an isogenic strain background. In order to create a solid epidemiological basis for assessing the actual relevance of dalbavancin resistance and its future development, we suggest that dalbavancin be routinely tested in clinically relevant MRSA isolates. On such a basis, further research projects could be planned to early detect MIC creeps early as well as to uncover and shed light on the herein discussed putative dalbavancin resistance mechanisms.
Data availability
The sequencing data is available at https://www.ncbi.nlm.nih.gov/ under accession numbers SRS21013891 for isolate 19-362 and SRS3676602 for isolate 16-33.
Abbreviations
- S.:
-
Staphylococcus
- MRSA:
-
Methicillin-resistant Staphylococcus aureus
- BSI:
-
Bloodstream infections
- MIC:
-
Minimal inhibitory concentrations
- TEM:
-
Transmission electron microscopy
- EUCAST:
-
European Committee on Antimicrobial Susceptibility Testing
- PBP2a:
-
Penicillin-binding protein 2a
- AST:
-
Antimicrobial susceptibility testing
- WGS:
-
Whole genome sequencing
- MHE:
-
Müller-Hinton E
- BMD:
-
Broth microdilution
- CA-MHB:
-
Cation-Adjusted Mueller-Hinton Broth
- MSSA:
-
Methicillin-susceptible Staphylococcus aureus
- SNV:
-
Single nucleotide variant
- MLST:
-
Multi locus sequence typing
References
Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–32.
Kourtis AP, Hatfield K, Baggs J, Mu Y, See I, Epson E, et al. Vital signs: Epidemiology and recent trends in Methicillin-resistant and in Methicillin-Susceptible Staphylococcus aureus Bloodstream Infections - United States. MMWR Morb Mortal Wkly Rep. 2019;68(9):214–9.
Mermel LA, Allon M, Bouza E, Craven DE, Flynn P, O’Grady NP, et al. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis. 2009;49(1):1–45.
The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. 2023;Version 13.1.
Dhand A, Sakoulas G. Reduced Vancomycin susceptibility among clinical Staphylococcus aureus isolates (‘the MIC Creep’): implications for therapy. F1000. Med Rep. 2012;4:4.
Périchon B, Courvalin P. VanA-type Vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53(11):4580–7.
Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175(1):117–27.
Andam CP, Fournier GP, Gogarten JP. Multilevel populations and the evolution of antibiotic resistance through horizontal gene transfer. FEMS Microbiol Rev. 2011;35(5):756–67.
Watkins RR, Lemonovich TL, File TM. Jr. An evidence-based review of linezolid for the treatment of methicillin-resistant Staphylococcus aureus (MRSA): place in therapy. Core Evid. 2012;7:131–43.
Garnock-Jones KP, Single-Dose Dalbavancin. A review in Acute bacterial skin and skin structure infections. Drugs. 2017;77(1):75–83.
Werth BJ, Ashford NK, Penewit K, Waalkes A, Holmes EA, Ross DH, et al. Dalbavancin exposure in vitro selects for dalbavancin-non-susceptible and Vancomycin-intermediate strains of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2021;27(6):910.e1-.e8.
Zhang R, Polenakovik H, Barreras Beltran IA, Waalkes A, Salipante SJ, Xu L, Werth BJ. Emergence of Dalbavancin, Vancomycin, and Daptomycin Nonsusceptible Staphylococcus aureus in a patient treated with dalbavancin: Case Report and isolate characterization. Clin Infect Dis. 2022;75(9):1641–4.
Jones RN, Flamm RK, Sader HS. Surveillance of dalbavancin potency and spectrum in the United States (2012). Diagn Microbiol Infect Dis. 2013;76(1):122–3.
Zhanel GG, Calic D, Schweizer F, Zelenitsky S, Adam H, Lagacé-Wiens PR, et al. New lipoglycopeptides: a comparative review of dalbavancin, oritavancin and telavancin. Drugs. 2010;70(7):859–86.
Streit JM, Fritsche TR, Sader HS, Jones RN. Worldwide assessment of dalbavancin activity and spectrum against over 6,000 clinical isolates. Diagn Microbiol Infect Dis. 2004;48(2):137–43.
Guzek A, Rybicki Z, Tomaszewski D. In vitro analysis of the minimal inhibitory concentration values of different generations of anti-methicillin-resistant Staphylococcus aureus antibiotics. Indian J Med Microbiol. 2018;36(1):119–20.
Koeth LM, DiFranco-Fisher JM, McCurdy S. A reference broth microdilution method for Dalbavancin in Vitro susceptibility testing of Bacteria that grow aerobically. J Vis Exp. 2015(103).
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011. 2011;17(1):3.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9.
Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–76.
Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes De Novo Assembler. Curr Protocols Bioinf. 2020;70(1):e102.
Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1):D517–25.
Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. 2014;52(5):1501–10.
Kussmann M, Karer M, Obermueller M, Schmidt K, Barousch W, Moser D, et al. Emergence of a dalbavancin induced glycopeptide/lipoglycopeptide non-susceptible Staphylococcus aureus during treatment of a cardiac device-related endocarditis. Emerg Microbes Infect. 2018;7(1):202.
Kuipers J, Giepmans BNG. Neodymium as an alternative contrast for uranium in electron microscopy. Histochem Cell Biol. 2020;153(4):271–7.
Jones RN, Sader HS, Flamm RK. Update of dalbavancin spectrum and potency in the USA: report from the SENTRY Antimicrobial Surveillance Program (2011). Diagn Microbiol Infect Dis. 2013;75(3):304–7.
Zhanel GG, Trapp S, Gin AS, DeCorby M, Lagacé-Wiens PR, Rubinstein E, et al. Dalbavancin and telavancin: novel lipoglycopeptides for the treatment of Gram-positive infections. Expert Rev Anti Infect Ther. 2008;6(1):67–81.
Ortwine JK, Werth BJ, Sakoulas G, Rybak MJ. Reduced glycopeptide and lipopeptide susceptibility in Staphylococcus aureus and the seesaw effect: taking advantage of the back door left open? Drug Resist Updat. 2013;16(3–5):73–9.
Werth BJ, Jain R, Hahn A, Cummings L, Weaver T, Waalkes A, et al. Emergence of dalbavancin non-susceptible, Vancomycin-intermediate Staphylococcus aureus (VISA) after treatment of MRSA central line-associated bloodstream infection with a dalbavancin- and Vancomycin-containing regimen. Clin Microbiol Infect. 2018;24(4):429.e1-.e5.
Njenga J, Nyasinga J, Munshi Z, Muraya A, Omuse G, Ngugi C, Revathi G. Genomic characterization of two community-acquired methicillin-resistant Staphylococcus aureus with novel sequence types in Kenya. Front Med (Lausanne). 2022;9:966283.
Howden BP, McEvoy CR, Allen DL, Chua K, Gao W, Harrison PF, et al. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog. 2011;7(11):e1002359.
Watanabe Y, Cui L, Katayama Y, Kozue K, Hiramatsu K. Impact of rpoB mutations on reduced Vancomycin susceptibility in Staphylococcus aureus. J Clin Microbiol. 2011;49(7):2680–4.
Wichelhaus TA, Böddinghaus B, Besier S, Schäfer V, Brade V, Ludwig A. Biological cost of rifampin resistance from the perspective of Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46(11):3381–5.
Wang Y, Li X, Jiang L, Han W, Xie X, Jin Y, et al. Novel Mutation sites in the development of Vancomycin- Intermediate Resistance in Staphylococcus aureus. Front Microbiol. 2016;7:2163.
Tanaka M, Onodera Y, Uchida Y, Sato K. Quinolone resistance mutations in the GrlB protein of Staphylococcus aureus. Antimicrob Agents Chemother. 1998;42(11):3044–6.
Conceição T, de Lencastre H, Aires-de-Sousa M. Prevalence of biocide resistance genes and chlorhexidine and mupirocin non-susceptibility in Portuguese hospitals during a 31-year period (1985–2016). J Glob Antimicrob Resist. 2021;24:169–74.
Bakthavatchalam YD, Babu P, Munusamy E, Dwarakanathan HT, Rupali P, Zervos M, et al. Genomic insights on heterogeneous resistance to Vancomycin and teicoplanin in Methicillin-resistant Staphylococcus aureus: a first report from South India. PLoS ONE. 2019;14(12):e0227009.
Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML. Reduced Vancomycin susceptibility in Staphylococcus aureus, including Vancomycin-intermediate and heterogeneous Vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev. 2010;23(1):99–139.
Fowler PW, Cole K, Gordon NC, Kearns AM, Llewelyn MJ, Peto TEA, et al. Robust prediction of resistance to Trimethoprim in Staphylococcus aureus. Cell Chem Biol. 2018;25(3):339–e494.
Wang T, Tanaka M, Sato K. Detection of grlA and gyrA mutations in 344 Staphylococcus aureus strains. Antimicrob Agents Chemother. 1998;42(2):236–40.
Jian Y, Lv H, Liu J, Huang Q, Liu Y, Liu Q, Li M. Dynamic changes of Staphylococcus aureus susceptibility to Vancomycin, Teicoplanin, and Linezolid in a Central Teaching Hospital in Shanghai, China, 2008–2018. Front Microbiol. 2020;11:908.
Shariati A, Dadashi M, Chegini Z, van Belkum A, Mirzaii M, Khoramrooz SS, Darban-Sarokhalil D. The global prevalence of Daptomycin, Tigecycline, Quinupristin/Dalfopristin, and linezolid-resistant Staphylococcus aureus and coagulase-negative staphylococci strains: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2020;9(1):56.
Chen YH, Liu CY, Ko WC, Liao CH, Lu PL, Huang CH, et al. Trends in the susceptibility of methicillin-resistant Staphylococcus aureus to nine antimicrobial agents, including ceftobiprole, nemonoxacin, and tyrothricin: results from the Tigecycline in Vitro Surveillance in Taiwan (TIST) study, 2006–2010. Eur J Clin Microbiol Infect Dis. 2014;33(2):233–9.
Acknowledgements
Not applicable.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Conceptualization, H.L., J.F.H., K.SR., R.K.; methodology, K.SR., R.K., K.SD., K.SL.; validation, K.SR., R.K., K.SP., P.S., K.ST.; formal analysis, J.F.H., K.SR., R.K., L.S., N.C., K.ST., investigation, J.F.H., M.S., K.SR., R.K., H.L.; resources, H.L.; data curation, J.F.H., M.S., K.SR., R.K.; writing—original draft preparation, J.F.H., M.S.; writing—review and editing, K.SR., R.K., L.S., K.SD., J.B.H., A.L., F.L., S.D., M.R., H.B., H.L.; visualization, J.F.H., M.S., K.SR., L.S.; supervision, H.L., R.K., J.B.H.; project administration, H.L.; All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki. It was part of the study “Epidemiology, resistance, virulence factors and clinic of Staphylococcaceae bacteremia in the General Hospital of the Medical University of Vienna” approved by the local ethics committee of the Medical University of Vienna (EC No.: 1316/2017). Since this was a retrospective analysis of routine blood cultures from the General Hospital of the Medical University of Vienna, and originally no samples were taken for scientific purposes, according to the Ethics Committee, no informed consent to participate was required.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hotz, J.F., Staudacher, M., Schefberger, K. et al. Unraveling novel mutation patterns and morphological variations in two dalbavancin-resistant MRSA strains in Austria using whole genome sequencing and transmission electron microscopy. BMC Infect Dis 24, 899 (2024). https://doi.org/10.1186/s12879-024-09797-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-024-09797-w