Abstract
Background
Brain network connectivity modeling is a crucial method for studying the brain’s cognitive functions. Meta-analyses can unearth reliable results from individual studies. Meta-analytic connectivity modeling is a connectivity analysis method based on regions of interest (ROIs) which showed that meta-analyses could be used to discover brain network connectivity.
Results
In this paper, we propose a new meta-analysis method that can be used to find network connectivity models based on the Apriori algorithm, which has the potential to derive brain network connectivity models from activation information in the literature, without requiring ROIs. This method first extracts activation information from experimental studies that use cognitive tasks of the same category, and then maps the activation information to corresponding brain areas by using the automatic anatomical label atlas, after which the activation rate of these brain areas is calculated. Finally, using these brain areas, a potential brain network connectivity model is calculated based on the Apriori algorithm. The present study used this method to conduct a mining analysis on the citations in a language review article by Price (Neuroimage 62(2):816–847, 2012). The results showed that the obtained network connectivity model was consistent with that reported by Price.
Conclusions
The proposed method is helpful to find brain network connectivity by mining the co-activation relationships among brain regions. Furthermore, results of the co-activation relationship analysis can be used as a priori knowledge for the corresponding dynamic causal modeling analysis, possibly achieving a significant dimension-reducing effect, thus increasing the efficiency of the dynamic causal modeling analysis.
Similar content being viewed by others
Background
Functional neuroimaging, especially functional magnetic resonance imaging (fMRI), is an indispensable method for non-invasively researching human brain function. fMRI is not only used to study the function of a particular brain region, but is also increasingly being used to establish the network structure of the brain. The human brain is comprised of highly complex networks with hundreds of billions of neurons and more than 100 brain regions [1]. The various brain regions work both independently and collaboratively to complete certain cognitive tasks. Over the last 20 years, many studies have investigated brain activation using fMRI. However, most of these studies only examined brain activation in response to a specific task; while we gained knowledge of discrete brain areas by those studies, we still lack information about the functional integration (connections) among them.
Meta-analysis is an increasingly popular and valuable tool for summarizing results across many neuroimaging studies. Currently, two meta-analysis methods are popular in the brain imaging literature: the activation likelihood estimation (ALE) meta-analysis method [2, 3] and the meta-analytic connectivity modeling (MACM) method [4]. The ALE meta-analysis method can effectively integrate studies with consistent results by using statistical analyses; however, ALE is unsuitable for modeling the existing functional connections in the brain. On the other hand, the MACM method is based on the BrainMap database [5], which examines the relationships and connections between a particular region of interest (ROI) and other ROIs. Furthermore, MACM can overlay the results of separate analyses of multiple ROIs to obtain the corresponding network connectivity model.
In this paper, we present a new meta-analysis method for mining the co-activation relationship of brain regions without using ROIs. Our method focuses on the functional brain connectivity under the same task. This method uses the automatic anatomical label (AAL) atlas to define the brain region of each foci reported, and then applies the Apriori algorithm [6] to calculate the co-activation relationships. To confirm the effectiveness of this method, we employed part of a literature review [7] containing a meta-analytic dataset and compared the results of our meta-analysis to those obtained in the literature review. Furthermore, the possible dimension-reducing effects of this method on the corresponding dynamic casual modeling (DCM) analysis were analyzed. This will enable a higher efficiency in DCM when analyzing the effective connectivity of multiple ROIs.
Methods
The proposed method aimed to find the co-activation relationships among brain regions from a dataset consisting of neuroimaging studies. The method includes three steps. First, all activation foci are assigned to the identified brain regions. Second, brain regions that frequently appear across the studies are identified using an association analysis. Finally, the associated network of related brain regions is calculated using the Apriori algorithm. The details of the new method are described below.
Our brain region activation probability model is based on the voxel activation probability model of the ALE method [2]. When modeling the voxel activation, we assumed that for a certain voxel coordinate Xi, the probability of being activated at the peak point (x, y, z) of a certain region is:
where di is the Euclidean distance from Xi to point (x, y, z), and σ is the standard deviation of the distribution.
For a point in the brain Xi, the overall probably that this point will be activated can be calculated as follows:
where a, b indicates a different peak coordinate for activation (note that if there are more than two peak coordinates, then formula (2) will change according to the inclusion–exclusion principle in probability).
Formula (2) can be used to obtain the probability of each point in the brain being activated in each study and the probability distribution, which are then represented as a brain activation probability distribution diagram. Then we can obtain the border of a brain region’s anatomy, V, by querying the anatomical boundaries found in various brain region annotated databases. Here, we used the AAL atlas. An integral was used to determine the boundary of the activated region. For a particular brain region, the possibility that it is active is defined as:
where, Pr (Xi) is the probability that point Xi is activated in region V, which is obtained in the first step, and Pr(V) is the probably that a certain region V is activated. The region is output by name when Pr(V) is greater than a certain threshold t, thus obtaining the activated anatomical region in the study. The finer the division of a brain region’s anatomical boundaries, the more accurate the calculation of the brain network.
The Apriori algorithm [6] mainly uses prior knowledge of the data to perform its analyses; therefore, it can take advantage of mining the frequent itemsets. Here, we adopted the A-Close algorithm [8] of the Apriori algorithm.
The A-Close algorithm also mainly uses prior knowledge, as follows: (1) a subset of all frequent itemsets is also a frequent itemset, (2) a superset of all of the non-frequent itemsets is also non-frequent, (3) the degree of support of a frequent itemset is equal to its closure, and (4) frequent itemsets only have one closure.
Here, we made the following provision: A is an exhaustive list of brain activation regions under a particular cognitive state; support (A) indicates the number of times that list A appears in several articles divided by the total number of articles. If support (A) is greater than a threshold value, such as 0.8, then it is termed a frequent itemset, and the threshold value is named a degree of support of A. The possibility of determining connections between brain regions from a particular brain origin is a very large inner frequent itemset. In a frequent itemset A, the confidence coefficient for deducing region b from brain region a is calculated as:
It is assumed that there is a connection from A to B if confidence (A → B) is greater than a threshold value, such as 0.7.
This method first identifies the sets of brain regions that are activated frequently in studies utilizing similar tasks. Then, it obtains the degree of mutual influence within each brain area by calculating the confidence coefficient among the brain areas in this set. Finally, it generates a co-activation relationship model of the brain for studies utilizing the same tasks.
To demonstrate our methods, we used the reference list within a recent review article [7] as our dataset; the review article we used discussed the brain areas associated with heard speech, speech production, and reading. The benefits of utilizing such experimental materials are two-fold. First, all of the studies used the same type of experiment, that is, experiments activating brain processes reflecting orthography mapping to phonology. This ensured that the experimental data were not affected by noise interference, which improved the accuracy of the analysis results. Second, as the experimental materials were collected from an authoritative review paper, we can verify the correctness of the algorithm by comparing the network reported by Price [7] with the network obtained by the automatic analysis.
The paper by Price [7] is divided into several sections, and the final section, “Dissociating neural pathways for mapping orthography to phonology,” discusses how orthography is mapped to phonology when the brain performs reading tasks. The process is generally divided into two stages, namely orthographic analysis and orthographic coding mapped to phonology. The author, through the inductive analysis of the review in the above-mentioned section, concludes that there is a brain network for completing the above two stages.
There are 62 studies mentioned in the “Dissociating neural pathways for mapping orthography to phonology” section of the review. Thirty-three studies (Table 1) that fulfilled the following criteria were included in our meta-analysis: (1) used fMRI or positron emission tomography as the imaging modality, (2) applied visual tasks that required speech processing, (3) used whole-brain scanning and reported complete coordinates of the activation in standardized stereotaxic space [the Talairach atlas or the Montreal Neurological Institute (MNI) atlas], and (4) recruited healthy subjects.
To find the co-activation relationships of the brain regions from the above dataset, we first extracted the activation information and converted it into a text file that included the peak coordinates (x, y, z). All of the coordinates were transferred to the corresponding MNI coordinates. Next, using formula (3), the brain coordinates were mapped to the corresponding brain regions in which the probability-activated threshold was defined as 0.9. Next, the A-Close algorithm was used to calculate the associated network of related brain regions, among them, the A-Close algorithm of minimum support was defined as 0.8 and the minimum confidence as 0.7. Finally, we used Pajek [42] to graph the experimental results (Pajek can be applied to analyze large-scale complex networks and is a powerful tool for studying a variety of complicated nonlinear networks).
Results and discussion
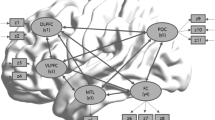
Our meta-analysis method examined how orthography is mapped to the functional connections related to phonology. The experimental results are shown in Fig. 1, wherein the weight value in red represents the confidence coefficient of each edge.
The weight values (red numbers) indicate the confidence coefficient of each edge (Frontal_Inf_Oper_L: left inferior frontal gyrus, opercular part; Frontal_Inf_Tri_L: left inferior frontal gyrus, triangular part; Frontal_Mid_L: left middle frontal gyrus; Fusiform_L: left fusiform gyrus; Occipital_Inf_L: left inferior occipital gyrus; Occipital_Mid_L: left middle occipital gyrus; Postcentral_L: left postcentral gyrus; Precentral_L: left precentral gyrus; Supp_Motor_Area_L: left supplementary motor area; Temporal_Inf_L: left inferior temporal gyrus).
As shown in Fig. 1, the left precentral gyrus, left fusiform, and left occipital lobe are connected, while the left temporal gyrus and fusiform gyrus are connected to the left precentral gyrus. There is one edge connection line between the left precentral gyrus and left inferior frontal gyrus. There is also one edge between the left middle frontal gyrus and left precentral gyrus and one between the left supplementary motor area and left precentral gyrus.
The left precentral gyrus is the hub for all of the connections, and the areas that are most frequently co-activated along with the left precentral gyrus are the left frontal lobe, left postcentral gyrus, left inferior occipital, and left supplementary motor area.
The results showed that the network extends throughout the frontal and occipital areas of the brain. The posterior network runs from the occipital lobe to the precentral gyrus and through the fusiform gyrus, superior temporal gyrus, and middle occipital gyrus, among others. The more anterior network includes the middle frontal gyrus, inferior frontal gyrus, and supplementary motor area. The entire network terminates at the precentral gyrus.
The occipital area in cognitive science is referred to as the visual cortex, and its main function is to process visual information [43]. Because reading involves transforming visual text into information that can be encoded in the brain, the functional network for reading likely involves the occipital area. Orthographic analysis is often performed in the fusiform gyrus region [44, 45]; in the present study, one edge existed between the left inferior occipital gyrus and left inferior temporal gyrus (Fig. 1), which is consistent with the results reported by Price [7]. The left inferior temporal gyrus is thought to be a memory storage area [46]. Therefore, after the orthographic analysis stage is complete, the fusiform is likely involved in the orthographic coding stage, while phonology processing occurs in the brain’s memory area. Finally, the circuit is completed in the posterior region of the brain.
Two pathways are activated simultaneously during reading [47, 48], namely the phonetic and semantic pathways. The frontal lobe is mainly involved in grammar processing, semantic analysis [49], and word generation; thus, the network in the anterior part of the brain consists of the entire inferior frontal gyrus (opercular, orbital, and triangular portions). Another part of the network, the middle frontal gyrus, mainly executes semantic retrieval [50]. Both silent reading and reading aloud require the precentral gyrus and posterior central gyrus, which are involved in speech output [51, 52]; therefore, the precentral gyrus may be the hub node and output node of the network.
Collectively, the co-activation relationships of the brain regions (regarding how orthography is mapped to phonology when performing reading tasks) obtained with our analysis method were in accordance with the network reported in the literature review by Price [7]. The main difference between our study and the review is that in addition to containing the brain networks derived from our meta-analysis of functional connections reported in the literature, the review also contains a dorsal pathway that includes the dorsal precentral gyrus and extends from the superior temporal gyrus to the inferior occipital lobe. However, in the summary by Price [7], it is unclear whether the dorsal pathway exists. Our meta-analysis method indicates that the dorsal pathway likely does not exist. The reason for this belief is based on the following principles of the association rule algorithm: (1) more than 20 % of the articles did not simultaneously refer to the mentioned brain regions in the dorsal pathway, and (2) if more than 80 % of the articles referred to the mentioned brain regions in the dorsal pathway, then the confidence coefficient among the brain regions in the dorsal pathway would be lower than 70 %. However, this remains to be confirmed.
Furthermore, the results of the connectivity analysis can be used as a priori knowledge for a DCM analysis [53, 54]; this would produce a significant dimension-reducing effect when performing the DCM analysis, thus increasing the efficiency of the analysis. In this study, some brain areas had no connections (such as the left inferior occipital gyrus and left middle frontal gyrus), which may not need to be considered in the corresponding DCM analysis. Therefore, when performing DCM analyses for multiple ROIs, the reduction in the calculation of a single edge will reduce the dimension in an exponential order, thus significantly increasing the DCM analysis efficiency. The related comparative validation will be performed in a future study.
The meta-analysis method presented herein can determine the co-activation relationships among the brain regions activated in studies utilizing similar tasks; however, our study also has some limitations. First, we selected a meta-analysis dataset from a review that may have included the author’s subjective views; in the future, we will apply this method to a literature database. Second, in this paper, we used the AAL atlas to define the brain regions. As more research on the brain is conducted, atlases that are more precise may become available. The use of such precise atlases will improve the accuracy of the experimental results.
Conclusions
This article proposes a new meta-analysis method to find brain network connection models. This method employs data mining to discover the co-activation relationships among brain regions based on activation data reported in the relevant literature. Furthermore, studies utilizing word-reading tasks reported in a review by Price [7] were analyzed and the feasibility and robustness of this method were verified. Our meta-analysis method may enable us to better understand the brain’s processing patterns during certain cognitive tasks. In particular, the results of the connectivity analysis can provide reliable a priori knowledge for future experimental research. For example, the results of the connectivity analysis can be used in corresponding DCM analyses to eliminate edges that do not need to be considered in order to increase the DCM calculation efficiency.
Abbreviations
- AAL:
-
automatic anatomical label
- ALE:
-
activation likelihood estimation
- DCM:
-
dynamic causal modeling
- fMRI:
-
functional magnetic resonance imaging
- MACM:
-
meta-analytic connectivity modeling
- MNI:
-
Montreal Neurological Institute
- ROIs:
-
regions of interest
References
Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10(3):186–98.
Turkeltaub PE, Eden GF, Jones KM, Zeffiro TA. Meta-analysis of the functional neuroanatomy of single-word reading: method and validation. Neuroimage. 2002;16(3):765–80.
Eickhoff SB, Laird AR, Grefkes C, Wang LE, Zilles K, Fox PT. Coordinate-based activation likelihood estimation meta-analysis of neuroimaging data: a random-effects approach based on empirical estimates of spatial uncertainty. Hum Brain Mapp. 2009;30(9):2907–26.
Robinson JL, Laird AR, Glahn DC, Lovallo WR, Fox PT. Metaanalytic connectivity modeling: delineating the functional connectivity of the human amygdala. Hum Brain Mapp. 2010;31(2):173–84.
Fox PT, Lancaster JL. Mapping context and content: the BrainMap model. Nat Rev Neurosci. 2002;3(4):319–21.
Agrawal R, Srikant R. Fast algorithms for mining association rules. In: Proceedings of 20th international conference on very large data bases, VLDB. 1994. p. 487–99.
Price CJ. A review and synthesis of the first 20 years of PET and fMRI studies of heard speech, spoken language and reading. Neuroimage. 2012;62(2):816–47.
Pasquier N, Bastide Y, Taouil R, Lakhal L. Discovering frequent closed itemsets for association rules. In: Proceedings of 7th international conference on database theory (ICDT-99), vol 1540; 1999. p. 398–416.
Binder JR, Medler DA, Desai R, Conant LL, Liebenthal E. Some neurophysiological constraints on models of word naming. Neuroimage. 2005;27:677–93.
Bolger DJ, Hornickel J, Cone NE, Burman DD, Booth JR. Neural correlates of orthographic and phonological consistency effects in children. Hum Brain Mapp. 2008;29:1416–29.
Bolger DJ, Minas J, Burman DD, Booth JR. Differential effects of orthographic and phonological consistency in cortex for children with and without reading impairment. Neuropsychologia. 2008;46:3210–24.
Cohen L, Dehaene S, Vinckier F, Jobert A, Montavont A. Reading normal and degraded words: contribution of the dorsal and ventral visual pathways. Neuroimage. 2008;40:353–66.
Dietz NAE, Jones KM, Gareau L, Zeffiro TA, Eden GF. Phonological decoding involves left posterior fusiform gyrus. Hum Brain Mapp. 2005;26:81–93.
Edwards JD, Pexman PM, Goodyear BG, Chambers CG. An fMRI investigation of strategies for word recognition. Cogn Brain Res. 2005;24:648–62.
Fiebach CJ, Friederici AD, Muller K, von Cramon DY. fMRI evidence for dual routes to the mental lexicon in visual word recognition. J Cogn Neurosci. 2002;14:11–23.
Frost SJ, Mencl WE, Sandak R, Moore DL, Rueckl JG, Katz L, Fulbright RK, Pugh KR. A functional magnetic resonance imaging study of the tradeoff between semantics and phonology in reading aloud. NeuroReport. 2005;16:621–4.
Graves WW, Binder JR, Desai RH, Conant LL, Seidenberg MS. Neural correlates of implicit and explicit combinatorial semantic processing. Neuroimage. 2010;53:638–46.
Hagoort P, Indefrey P, Brown C, Herzog H, Steinmetz H, Seitz RJ. The neural circuitry involved in the reading of German words and pseudowords: a PET study. J Cogn Neurosci. 1999;11:383–98.
Herbster AN, Mintun MA, Nebes RD, Becker JT. Regional cerebral blood flow during word and nonword reading. Hum Brain Mapp. 1997;5:84–92.
Hu W, Lee HL, Zhang Q, Liu T, Geng LB, Seghier ML, Shakeshaft C, Twomey T, Green DW, Yang YM, Price CJ. Developmental dyslexia in Chinese and English populations: dissociating the effect of dyslexia from language differences. Brain. 2010;133:1694–706.
Ino T, Nakai R, Azuma T, Kimura T, Fukuyama H. Recognition and reading aloud of kana and kanji word: an fMRI study. Brain Res Bull. 2009;78:232–9.
Lee CY, Tsai JL, Kuo WJ, Yeh TC, Wu YT, Ho LT, Hung DL, Tzeng OJL, Hsieh JC. Neuronal correlates of consistency and frequency effects on Chinese character naming: an event-related MRI study. Neuroimage. 2004;23:1235–45.
Levy J, Pernet C, Treserras S, Boulanouar K, Berry I, Aubry F, Demonet J-F, Celsis P. Piecemeal recruitment of left-lateralized brain areas during reading: a spatio-functional account. NeuroImage. 2008;43:581–91.
Liu L, Deng X, Peng D, Cao F, Ding G, Jin Z, Zeng Y, Li K, Zhu L, Fan N, Deng Y, Bolger DJ, Booth JR. Modality- and task-specific brain regions involved in Chinese lexical processing. J Cogn Neurosci. 2009;21:1473–87.
Matsuo K, Chen S-HA, Hue C-W, Wu C-Y, Bagarinao E, Tseng W-YI, Nakai T. Neural substrates of phonological selection for Japanese character Kanji based on fMRI investigations. Neuroimage. 2010;50:1280–91.
McDermott KB, Petersen SE, Watson JM, Ojemann JG. A procedure for identifying regions preferentially activated by attention to semantic and phonological relations using functional magnetic resonance imaging. Neuropsychologia. 2003;41:293–303.
Mechelli A, Friston KJ, Price CJ. The effects of presentation rate during word and pseudoword reading: a comparison of PET and fMRI. J Cogn Neurosci. 2000;12:145–56.
Mechelli A, Gorno-Tempini ML, Price CJ. Neuroimaging studies of word and pseudoword reading: consistencies, inconsistencies, and limitations. J Cogn Neurosci. 2003;15:260–71.
Mechelli A, Crinion JT, Long S, Friston KJ, Ralph MAL, Patterson K, McClelland JL, Price CJ. Dissociating reading processes on the basis of neuronal interactions. J Cogn Neurosci. 2005;17:1753–65.
Meschyan G, Hemandez AE. Impact of language proficiency and orthographic transparency on bilingual word reading: an fMRI investigation. Neuroimage. 2006;29:1135–40.
Nosarti C, Mechelli A, Green DW, Price CJ. The impact of second language learning on semantic and nonsemantic first language reading. Cereb Cortex. 2010;20:315–27.
Owen WJ, Borowsky R, Sarty GE. FMRI of two measures of phonological processing in visual word recognition: ecological validity matters. Brain Lang. 2004;90:40–6.
Price CJ, McCrory E, Noppeney U, Mechelli A, Moore CJ, Biggio N, Devlin JT. How reading differs from object naming at the neuronal level. Neuroimage. 2006;29:643–8.
Rapp B, Lipka K. The literate brain: the relationship between spelling and reading. J Cogn Neurosci. 2011;23:1180–97.
Rumsey JM, Horwitz B, Donohue BC, Nace K, Maisog JM, Andreason P. Phonological and orthographic components of word recognition—a PET-rCBF study. Brain. 1997;120:739–59.
Seghier ML, Lee HL, Schofield T, Ellis CL, Price CJ. Inter-subject variability in the use of two different neuronal networks for reading aloud familiar words. NeuroImage. 2008;42:1226–36.
Tan LH, Feng CM, Fox PT, Gao JH. An fMRI study with written Chinese. NeuroReport. 2001;12:83–8.
Tan LH, Liu HL, Perfetti CA, Spinks JA, Fox PT, Gao JH. The neural system underlying Chinese logograph reading. NeuroImage. 2001;13:836–46.
Vigneau M, Jobard G, Mazoyer B, Tzourio-Mazoyer N. Word and non-word reading: what role for the visual word form area? Neuroimage. 2005;27:694–705.
Xiao ZW, Zhang JX, Wang XY, Wu RH, Hu XP, Weng XC, Tan LH. Differential activity in left inferior frontal gyrus for pseudowords and real words: an event-related fMRI study on auditory lexical decision. Hum Brain Mapp. 2005;25:212–21.
Xu B, Grafman J, Gaillard WD, Ishii K, Vega-Bermudez F, Pietrini P, Reeves-Tyer P, DiCamillo P, Theodore W. Conjoint and extended neural networks for the computation of speech codes: the neural basis of selective impairment in reading words and pseudowords. Cereb Cortex. 2001;11:267–77.
Batagelj V, Mrvar A. Pajek-analysis and visualization of large networks. In: Junger M, Mutzel P, editors. Graph drawing software, mathematics and visualization. Berlin, Heidelberg: Springer; 2004. p. 77–103. doi:10.1007/978-3-642-18638-74.
Zeki S, Watson JDG, Lueck CJ, Friston KJ, Kennard C, Frackowiak RSJ. A direct demonstration of functional specialization in human visual-cortex. J Neurosci. 1991;11(3):641–9.
Dehaene S, Cohen L. The unique role of the visual word form area in reading. Trends Cognit Sci. 2011;15(6):254–62.
Szwed M, Dehaene S, Kleinschmidt A, Eger E, Valabregue R, Amadon A, Cohen L. Specialization for written words over objects in the visual cortex. Neuroimage. 2011;56(1):330–44.
Creem SH, Proffitt DR. Defining the cortical visual systems: “what”, “where”, and “how”. Acta Psychol. 2001;107(1):43–68.
Hickok G, Poeppel D. Dorsal and ventral streams: a framework for understanding aspects of the functional anatomy of language. Cognition. 2004;92(1–2):67–99.
Hickok G, Poeppel D. Opinion—the cortical organization of speech processing. Nat Rev Neurosci. 2007;8(5):393–402.
Peelle JE, McMillan C, Moore P, Grossman M, Wingfield A. Dissociable patterns of brain activity during comprehension of rapid and syntactically complex speech: evidence from fMRI. Brain Lang. 2004;91(3):315–25.
McCarthy G, Blamire AM, Rothman DL, Gruetter R, Shulman RG. Echo-planar magnetic-resonance-imaging studies of frontal-cortex activation during word generation in humans. Proc Natl Acad Sci USA. 1993;90(11):4952–6.
Booth JR, Lu D, Burman DD, Chou TL, Jin Z, Peng DL, Zhang L, Ding GS, Deng Y, Liu L. Specialization of phonological and semantic processing in Chinese word reading. Brain Res. 2006;1071(1):197–207.
Price CJ, Mechelli A. Reading and reading disturbance. Curr Opin Neurobiol. 2005;15(2):231–8.
Friston KJ, Harrison L, Penny W. Dynamic causal modelling. NeuroImage. 2003;19(4):1273–302.
Friston KJ, Li B, Daunizeau J, Stephan KE. Network discovery with DCM. NeuroImage. 2011;56(3):1202–21.
Authors’ contributions
ZN and YN conceived of the study. ZN, YN, and QZ developed the algorithms. YN and LZ designed the experiment. LZ and JW performed the experiments and analyzed the data. ZN, YN, and QZ wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This research was supported by the National Strategic Basic Research Program (“973” Program) of the Ministry of Science and Technology of China (Grant Number: 2012CB7207002).
Availability of data and material
This paper’s meta-analysis data come from published literature.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Niu, Z., Nie, Y., Zhou, Q. et al. A brain-region-based meta-analysis method utilizing the Apriori algorithm. BMC Neurosci 17, 23 (2016). https://doi.org/10.1186/s12868-016-0257-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12868-016-0257-8