Abstract
Background
Culturomics can ascertain traces of microorganisms to be cultivated using different strategies and identified by matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry or 16S rDNA sequencing. However, to cater to all requirements of microorganisms and isolate as many species as possible, multiple culture conditions must be used, imposing a heavy workload. In addition, the fast-growing bacteria (e.g., Escherichia) surpass the slow-growing bacteria in culture by occupying space and using up nutrients. Besides, some bacteria (e.g., Pseudomonas) suppress others by secreting antibacterial metabolites, making it difficult to isolate bacteria with lower competence. Applying inhibitors to restrain fast-growing bacteria is one method to cultivate more bacterial species from human feces.
Results
We applied CHIR-090, an LpxC enzyme inhibitor that has antibacterial activity against most Gram-negative bacteria, to culturomics of human fresh feces. The antibacterial activity of CHIR-090 was first assessed on five Gram-negative species of bacteria (Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, Proteus vulgaris, and Bacteroides vulgatus), all of which are commonly isolated from the human gut. Then, we assessed suitable concentrations of the inhibitor. Finally, CHIR-090 was applied in blood culture bottles for bacterial cultivation. In total, 102 species from five samples were identified. Of these, we found one new species, two species not reported previously in the human gut, and 11 species not previously isolated from humans.
Conclusions
CHIR-090 can suppress E. coli, P. aeruginosa, K. pneumoniae, Pro. vulgaris, but not B. vulgatus. Compared with the non-inhibitor group, CHIR-090 increased bacteria isolation by 23.50%, including four species not reported in humans and one new species. Application of LpxC enzyme inhibitor in culturomics increased the number of species isolated from the human gut.
Similar content being viewed by others
Background
Recent studies have revealed that the gut microbiota plays an important role in maintaining homeostasis and human health [1, 2]. The revolution of metagenomics has helped further our comprehension of the human gut microbiota, but it has left a large number of unidentified sequences that may belong to unknown species [3, 4]. The renaissance of culturomics is trying to fill these gaps by isolating and analyzing the pure culture of human gut microorganisms [5, 6]. To date, more than 1000 species of bacteria in the human gut have been cultivated and studied [7]. However, because culturomics requires multiple cultivation conditions to satisfy the preferences of different species in the microbiota, it is a time- and labor-consuming process, which has long hindered progress in this area [8]. In our work isolating gut microorganisms, we found that Escherichia coli, a rapidly and easily grown species, quickly occupied most of the space of a culture plate, making it more difficult to isolate other bacterial species. In addition, the culture diversity of fecal samples with large numbers of Pseudomonas aeruginosa is much lower than that of samples without P. aeruginosa. Therefore, we postulate that inhibiting the growth of E. coli and P. aeruginosa in a culture system could result in a more efficient isolation of previously unidentified species.
CHIR-090 is an inhibitor of the enzyme LpxC, the key enzyme of lipid A biosynthesis in Gram-negative bacteria [9]. When lipid A biosynthesis, a cytoderm lipid that shields Gram-negative bacteria, is suppressed, the cytomembrane of Gram-negative bacteria can easily crack, leading to the death of bacteria [10]. Previous studies have demonstrated that CHIR-090 shows substantial antibacterial activity against both E. coli and P. aeruginosa [11].
In this study, we evaluated the antibacterial or inhibitory capacity of CHIR-090 against five Gram-negative bacteria. Furthermore, we determined the optimal concentration of CHIR-090 in blood culture bottles for fecal microbiota enrichment before prolonged cultivation. Finally, we applied CHIR-090 in culturomics of five fecal samples from healthy adults to evaluate the feasibility of using CHIR-090 to increase isolation of previously unreported bacterial species.
Results
Antibacterial capacity of CHIR-090
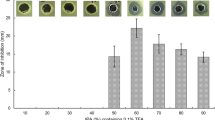
To evaluate the ability of CHIR-090 to suppress the growth of five Gram-negative bacteria (E. coli, P. aeruginosa, K. pneumoniae, Pro. vulgaris, and B. vulgatus), CHIR-090 was added to culture medium. We found that not all the Gram-negative bacteria were inhibited by CHIR-090 and the effective concentrations differed by species (Table 1). E. coli, P. aeruginosa, K. pneumoniae, and Pro. vulgaris were completely suppressed by CHIR-090 but at different concentrations of the inhibitor. Growth of Pro. vulgaris was inhibited with 8 μg/mL CHIR-090, whereas growth of E. coli and P. aeruginosa was inhibited at 40 μg/mL CHIR-090. K. pneumoniae was not inhibited until the concentration of CHIR-090 reached 200 μg/mL. Finally, growth of B. vulgatus was not affected by CHIR-090, even at the highest concentration tested. These differences in antibacterial activity may be related to different coding sequences (Additional file 1: Table S1) of enzyme LpxC, which result in diverse structures of this enzyme that affect the tightness of the binding with CHIR-090 [12].
Determination of optimal concentration of CHIR-090
In this study, we adopt Lagier’s group strategy [8]: we enriched fecal samples in blood culture bottles with 5% sheep blood and 5% rumen fluid and then subcultured the mixtures on YCFA plates, followed by subculture and identification of colonies. Although 200 μg/mL CHIR-090 could suppress 4 common fast-growing Gram-negative bacteria, we also tested CHIR-090 at 400 and 800 μg/mLl, because of the large numbers of microorganisms that the human gut harbors and the continuous consumption of the inhibitor during the prolonged enrichment of bacteria in our study. Because we observed that the effective concentration of CHIR-090 varied with bacterial species, we determined the optimal concentration before extending the experiment. To do so, fecal sample 1 (F1) was cultivated with CHIR-090 at 80, 400, and 800 μg/mL in blood culture bottles with 5% sheep blood and 5% rumen fluid; bottles with dimethyl sulphoxide (DMSO) and without treatment (blank) were used as controls. In this experiment, we isolated 19 species of bacteria in total. The percentages of colony-forming units of each bacterium under different CHIR-090 concentrations are shown in Additional file 1: Figure S1a, and that in anaerobic or aerobic conditions are shown in Additional file 1: Figure S1b and Additional file 1: Figure S1c, respectively. E. coli and Enterococcus faecium occupied the largest percentages in DMSO and blank bottles, in which only 6 and 5 species, respectively, were isolated. In the CHIR-090 bottles, growth of E. coli was suppressed, allowing the number of isolated species to increase to 12 species in the 80 and 400 μg/mL bottles, at equal percentages. However, when the concentration of CHIR-090 was increased to 800 μg/mL, the diversity decreased to 6 species; at 800 μg/mL, Enterococcus faecalis prevailed though no E. coli were found. Because CHIR-090 is a time-dependent inhibitor, we determined 400 μg/mL to be the optimal concentration for subsequent experiments.
Evaluation of CHIR-090 in human gut culturomics
We enriched fecal samples for 1 month, plating subsamples at 1, 3, 6, 12, 21, and 30 days, subculturing the resulting colonies in YCFA liquid medium, streak-inoculating the subcultures, and finally identifying the species by Matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI–TOF MS) or 16S rDNA sequencing (Fig. 1). Overall, we identified 102 species from five fresh fecal samples (Fig. 2 a). Taxonomic information showed that the isolates covered five phyla: Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Proteobacteria (Additional file 2: Table S2). Forty species were found only in 1 sample, 26 species were found in 2 samples, 14 species were found in 3 samples, 9 species were found in 4 samples, and 13 species were found in 5 samples (Fig. 2 b). The amount of overlap among samples is shown in Fig. 2 c. During the 1-month culture, different bacteria existed in different groups at different time points (Fig. 3 a), demonstrating that a prolonged culture time results in more complete isolation of species. The addition of CHIR-090 or not resulted in very different microecology, such that 23.5% of bacteria were only isolated in CHIR-090 bottles, 25.5% were only isolated from bottles without CHIR-090, and 51% were found in both bottles in general condition (including anaerobic and aerobic condition) (Fig. 3 b). Analysis of anaerobic or aerobic conditions respectively resulted in similar findings (Additional file 1: Figure S2a, Additional file 1: Figure S2b). The distributions of each species and percentages of each group in samples are shown in Additional file 1: Figure S3a-t. Moreover, addition of CHIR-090 accounted for about one-third of the bacterial isolates from one stool sample (Table 2), indicating that application of CHIR-090 results in discovery of more bacterial species and, to some degree, offsets the sample source deficiency. Among 102 species identified in our experiments, one isolate was a potentially novel species, two were not previously reported to be associated with the human gut, and 11 were not previously reported to be isolated from humans, four of which were from bottles with CHIR-090. The potentially novel bacterial species was also identified in a CHIR-090 bottle (Table 3). Eight of these species came from one sample and CHIR-090 accounted for five of them (Table 3).
Workflow of the culturomics strategy. Fecal samples were enriched in blood culture bottles containing 5% sheep blood, 5% rumen fluid, and CHIR-090 for one month, plating subsamples at 1, 3, 6, 12, 21, and 30 days, subculturing the resulting colonies in YCFA liquid medium (all cultures were preserved in glycerin at − 80 °C or liquid nitrogen for the reuse of bacteria), streak-inoculating the subcultures, and finally identifying the species by MALDI–TOF MS or 16S rDNA sequencing
Distribution and percentages of bacteria in each group. a The distribution of 102 bacteria in different groups at 6 points in time under general condition. b The percentage of all bacteria identified in blank bottles and CHIR-090 bottles under general, anaerobic, and aerobic conditions. Blue represents the blank group, yellow represents the CHIR-090 group, and green indicates presence in both bottles (common group)
Discussion
In recent decades, researchers have attempted to understand how the gut microbiome affects human health because it is the largest immune organ in the body [25,26,27,28]. Revolutions in sequencing techniques have added, little by little, to the landscape of the gut microbiome [29, 30]. However, this process has reached a plateau because sequencing techniques are limited in that they can only identify bacteria to the species level or, worse, can result in mismatched sequences [31]. Culturomics can identify bacteria to the strain level by applying multiple cultivation conditions to isolate the full diversity of the microbiota and by using MALDI–TOF MS or 16S rRNA amplification and sequencing for identification [32]. The isolates can be used in mechanistic studies, especially those that focus on interactions with other bacteria and with the host [33,34,35]. However, culturomics has its own limitations. First, to expand the culturable gut microbiota repertoire, complex conditions are essential to meet the various preferences of all species that make up the microbiota, which requires a great amount of work, although some studies have tried to simplify culture conditions [8]. This time- and labor-consuming workload has long hindered progress in culturomics. Second, interactions among bacteria increase the difficulty of isolation. For example, before slow-growing bacteria can grow sufficiently to be identified, E. coli have already dominated the culture and prevented further growth of lagging species. Moreover, an antibacterial peptide produced by Pseudomonas can affect the growth of other bacteria in co-culture [36]. Selected culture media can help distinguish bacteria, but selective media are generally used to isolate specific species and thus are not efficient for mass isolation [37, 38]. The use of phage is an option to kill Escherichia and Pseudomonas; however, the extreme specificity of phage to the strain level makes it a less effective option for stopping growth of various Escherichia and Pseudomonas species [39]. The use of antibacterial agents is another way to suppress fast-growing bacteria. CHIR-090 inhibits the enzyme LpxC, which catalyzes the first irreversible step of lipid A biosynthesis of E. coli and most Gram-negative bacteria, and aroused our interest [12]. Previous studies showed that CHIR-090 could inhibit the growth of E. coli and P. aeruginosa [9, 40]. In our studies, CHIR-090 excelled in suppressing growth of most Gram-negative bacteria that we studied, especially E. coli. Its ability to do so depends to a certain extent on the LpxC coding sequence, which determines the structure of LpxC. LpxC source of coding sequence on line may predict the effect of CHIR-090 before using it as an antibacterial agent [12].
Cultures with sheep blood and rumen fluid can satisfy a large majority of gut microorganisms [8]. In our previous studies (unpublished data) we found that prolonging the culture time can isolate more bacteria because different bacteria exist at different times over a month. We also found that more bacteria are isolated early in the 1-month culture period; however, after day 10, the microbiota varies less. Prolonging enrichment over 30 days is not recommended. Therefore, we chose to subculture on days 1, 3, 6, 12, 21, and 30. Fecal culture represents a complicated microbial ecology in that it varies throughout a 1-month enrichment. CHIR-090 is the main factor affecting microbial ecology because it inhibits growth of many of the Gram-negative bacteria (40% of Gram-negative bacteria are inhibited in this study), thus disturbing the microbiota. Meanwhile, it allows other bacteria to grow because space and nutrients are spared. Thus, without altering other conditions and adding CHIR-090 into blood culture bottles, the number of species isolated can be increased by at least a quarter. Increasing the number of samples would allow isolation of more bacteria; however, when a source is difficult to obtain, CHIR-090 can, to some degree, offset this deficiency. Therefore, we believe that CHIR-090 has great potential for isolating diverse species in the gut microbiota and discovering new bacterial species.
Conclusion
In this study, we introduced CHIR-090 in culturomics of human gut microbiota. First, we found that CHIR-090 could inhibit E. coli, P. aeruginosa, K. pneumoniae, and Pro. vulgaris,which are all commonly isolated species in human feces. Second we optimize the concentration of CHIR-090 in blood culture bottles for human feces culturing. Under the concentration of 400 μg/mL, CHIR-090 increased bacterial diverisity of isolates in five fecal samples by 23.50%, showing its usefulness in fecal microbiota culturomics. Application of LpxC enzyme inhibitor increased the number of species isolated from the human gut.
Methods
Bacterial strains
E. coli, P. aeruginosa, K. pneumoniae, Pro. vulgaris, and B. vulgatus were isolated from the human feces and preserved at − 80 °C in our laboratory. MALDI–TOF MS was conducted by QuantiHealth Technology Co. Ltd. (Beijing, China) to confirm the species identity of the inoculated colonies before usage.
CHIR-090
CHIR-090 (Beijing BioRab Technology Co. Ltd., Beijing, China), also called benzamide, N-[(1S,2R)-2-hydroxy-1-[(hydroxyamino)carbonyl] propyl]-4-[2-[4- (4-morpholinylmethyl) phenyl]ethynyl], is a two-step, slow, tight-binding inhibitor of E. coli LpxC. Before usage, CHIR-090 was dissolved in DMSO.
Antibacterial capacity of CHIR-090
Colonies of E. coli, P. aeruginosa, K. pneumoniae, Pro. vulgaris, and B. vulgatus were inoculated into YCFA liquid medium [41] with different concentrations of CHIR-090 (8, 40, 80, and 200 μg/mL). For controls, we inoculated colonies into bottles with DMSO and into bottles without any treatment (“blanks”). All bottles were incubated at 37 °C under aerobic conditions for 24 h. The cultures were then cultivated using the agar dilution method at 37 °C under aerobic conditions for 24 h. Finally, colony-forming units (CFU) on each agar plate were counted.
Stool samples and pretreatment
Six fresh fecal samples were collected from six healthy human adults who met the screening criteria for donors in the European Fecal Microbiota Transplantation (FMT) criteria [42] and were designated F1 to F6. Each fecal sample (500 mg) was diluted in 15 ml of sterile phosphate buffer saline (PBS) immediately after collection and blended thoroughly. Then, 0.5 ml of each sample suspension was injected into a blood culture bottle with 5% sheep blood and 5% rumen liquid for enrichment.
Optimization of the applied concentration of CHIR-090
Dissolved CHIR-090 was injected into blood culture bottles (with 5% sheep blood and 5% rumen fluid) to reach a concentration of 80, 400, or 800 μg/ml; bottles with DMSO and without treatments (blanks) were used as controls. After diluting 500 mg of fresh fecal sample F1 with 15 ml of sterile PBS, 0.5 ml of the diluted suspension was added into each prepared bottle (liquid volume 50 ml) and incubated at 37 °C, under anaerobic or aerobic condition for 24 h. The cultures were then cultivated using the agar dilution method at 37 °C under aerobic conditions for 24 h and anaerobic conditions for 48 h. The colonies were collected and identified by MALDI–TOF MS; if colonies were not identifiable, they were subjected to 16S rDNA sequencing.
Cultivation strategy
Blood cultures containing 5% sheep blood, 5% rumen fluid, and CHIR-090 (400 μg/ml) were used to enrich the fresh stool dilutions at 37 °C under anaerobic and aerobic conditions for 1 month. On days 1, 3, 6, 12, 21, and 30, samples of enriched cultures were extracted from the bottles by syringe, and doubling dilutions were spread onto YCFA plates for culture at 37 °C under aerobic conditions for 24 h or anaerobic conditions for 48 h. Colonies were picked according to their appearance (size, color, and shape) for subculture in YCFA liquid medium. The subcultures were then streak-inoculated and later identified by MALDI–TOF MS or 16S rDNA sequencing.
MALDI–TOF MS
Colonies were first identified by MALDI–TOF MS using an Autof ms1000 system (QuantiHealth Technology Co. Ltd., Beijing, China) after being deposited into 1 ml of lysis buffer (70% formic acid) and 1 ml of matrix solution (saturated α-cyano acid-4-hydroxycinnamic in 50% acetonitrile and 2.5% trifluoroacetic acid). Each spectrum was compared with those of known samples in the database. A colony was not labeled as credible at the species level without a total score ≥ 9.0.
16S rDNA sequencing
Colonies that were not identified by MALDI-TOF MS were subjected to 16S rDNA sequencing with primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (Tsingke Biological Technology Co. Ltd., Beijing, China). For identification at the species level, we chose a threshold similarity of > 98.0%. An isolate with a similarity value below this threshold was suspected to be a new species.
Classification of cultivated species
We used an online database of isolated bacteria in humans (http://hpr.mediterranee-infection.com/arkotheque/client/ihu_bacteries/recherche/index.php) to classify all isolates into four categories: new species, known species in human gut, species previously isolated from the environment but first isolated from humans, and species previously isolated from humans but first isolated from human gut. We also conducted literature searches on PubMed to compare against published papers and confirm the classification.
Availability of data and materials
All data generated or analyzed during current study are available from the corresponding author on reasonable request.
Abbreviations
- B. vulgatus :
-
Bacteroides vulgatus
- CFU :
-
Colony-forming units
- DMSO :
-
Dimethyl sulphoxide
- E. coli :
-
Escherichia coli
- FMT :
-
Fecal microbiota transplantation
- K. pneumoniare :
-
Klebsiella pneumoniae
- MALDI–TOF MS :
-
Matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry
- P. aeruginosa :
-
Pseudomonas aeruginosa
- PBS :
-
Phosphate buffer saline
- Pro. vulgaris :
-
Proteus vulgaris
References
Sommer F, Backhed F. The gut microbiota--masters of host development and physiology. NAT REV MICROBIOL. 2013;11(4):227–38.
Postler TS, Ghosh S. Understanding the Holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab. 2017;26(1):110–30.
Lepage P, Leclerc MC, Joossens M, Mondot S, Blottiere HM, Raes J, Ehrlich D, Dore J. A metagenomic insight into our gut's microbiome. GUT. 2013;62(1):146–58.
Lagier JC, Dubourg G, Million M, Cadoret F, Bilen M, Fenollar F, Levasseur A, Rolain JM, Fournier PE, Raoult D. Culturing the human microbiota and culturomics. NAT REV MICROBIOL. 2018:540–50.
Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, Goulding D, Lawley TD. Culturing of 'unculturable' human microbiota reveals novel taxa and extensive sporulation. NATURE. 2016;533(7604):543–6.
Lau JT, Whelan FJ, Herath I, Lee CH, Collins SM, Bercik P, Surette MG. Capturing the diversity of the human gut microbiota through culture-enriched molecular profiling. GENOME MED. 2016;8(1):72.
Rajilic-Stojanovic M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38(5):996–1047.
Lagier JC, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, Caputo A, Cadoret F, Traore SI, Seck EH, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203.
Barb AW, McClerren AL, Snehelatha K, Reynolds CM, Zhou P, Raetz CR. Inhibition of lipid a biosynthesis as the primary mechanism of CHIR-090 antibiotic activity in Escherichia coli. BIOCHEMISTRY-US. 2007;46(12):3793–802.
Kalinin DV, Holl R. Insights into the zinc-dependent Deacetylase LpxC: biochemical properties and inhibitor design. Curr Top Med Chem. 2016;16(21):2379–430.
McClerren AL, Endsley S, Bowman JL, Andersen NH, Guan Z, Rudolph J, Raetz CR. A slow, tight-binding inhibitor of the zinc-dependent deacetylase LpxC of lipid a biosynthesis with antibiotic activity comparable to ciprofloxacin. BIOCHEMISTRY-US. 2005;44(50):16574–83.
Barb AW, Jiang L, Raetz CR, Zhou P. Structure of the deacetylase LpxC bound to the antibiotic CHIR-090: time-dependent inhibition and specificity in ligand binding. Proc Natl Acad Sci U S A. 2007;104(47):18433–8.
Shivaji S, Chaturvedi P, Suresh K, Reddy GS, Dutt CB, Wainwright M, Narlikar JV, Bhargava PM. Bacillus aerius sp. nov., Bacillus aerophilus sp. nov., Bacillus stratosphericus sp. nov. and Bacillus altitudinis sp. nov., isolated from cryogenic tubes used for collecting air samples from high altitudes. Int J Syst Evol Microbiol. 2006;56(Pt 7):1465–73.
Ouoba L, Vouidibio MA, Thorsen L, Anyogu A, Nielsen DS, Kobawila SC, Sutherland JP. Lysinibacillus louembei sp. nov., a spore-forming bacterium isolated from Ntoba Mbodi, alkaline fermented leaves of cassava from the Republic of the Congo. Int J Syst Evol Microbiol. 2015;65(11):4256–62.
Shafi S, Kamili AN, Shah MA, Bandh SA, Dar R. Dynamics of bacterial class bacilli in the deepest valley lake of Kashmir-the Manasbal Lake. Microb Pathog. 2017;104:78–83.
Stevens MJ, Inglin RC, Meile L. Complete and Assembled Genome Sequence of Vagococcus teuberi DSM 21459T, a Novel Species Isolated from Fermented Cow Milk in Mali. Genome Announc. 2017;5(4).
Raats D, Halpern M. Oceanobacillus chironomi sp. nov., a halotolerant and facultatively alkaliphilic species isolated from a chironomid egg mass. Int J Syst Evol Microbiol. 2007;57(Pt 2):255–9.
Glaeser SP, McInroy JA, Busse HJ, Kampfer P. Pseudogracilibacillus auburnensis gen. Nov., sp. nov., isolated from the rhizosphere of Zea mays. Int J Syst Evol Microbiol. 2014;64(Pt 7):2442–8.
Chou JH, Chou YJ, Lin KY, Sheu SY, Sheu DS, Arun AB, Young CC, Chen WM. Paenibacillus fonticola sp. nov., isolated from a warm spring. Int J Syst Evol Microbiol. 2007;57(Pt 6):1346–50.
Fruhling A, Schumann P, Hippe H, Straubler B, Stackebrandt E. Exiguobacterium undae sp. nov. and Exiguobacterium antarcticum sp. nov. Int J Syst Evol Microbiol. 2002;52(Pt 4):1171–6.
Coorevits A, Dinsdale AE, Heyrman J, Schumann P, Van Landschoot A, Logan NA, De Vos P. Lysinibacillus macroides sp. nov., nom. Rev. Int J Syst Evol Microbiol. 2012;62(Pt 5):1121–7.
Satomi M, La Duc MT, Venkateswaran K. Bacillus safensis sp. nov., isolated from spacecraft and assembly-facility surfaces. Int J Syst Evol Microbiol. 2006;56(Pt 8):1735–40.
Adriaans B, Garelick H. Cytotoxicity of Fusobacterium ulcerans. J Med Microbiol. 1989;29(3):177–80.
Bollet C, Elkouby A, Pietri P, de Micco P. Isolation of Enterobacter amnigenus from a heart transplant recipient. Eur J Clin Microbiol Infect Dis. 1991;10(12):1071–3.
Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. NATURE. 2016;535(7610):75–84.
Brown EM, Kenny DJ, Xavier RJ. Gut microbiota regulation of T cells during inflammation and autoimmunity. Annu Rev Immunol. 2019;37:599–624.
Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. NAT REV IMMUNOL. 2013;13(5):321–35.
Liu H, Hu C, Zhang X, Jia W. Role of gut microbiota, bile acids and their cross-talk in the effects of bariatric surgery on obesity and type 2 diabetes. J Diabetes Investig. 2018;9(1):13–20.
Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM, et al. The gut microbiota and host health: a new clinical frontier. GUT. 2016;65(2):330–9.
Rojo D, Mendez-Garcia C, Raczkowska BA, Bargiela R, Moya A, Ferrer M, Barbas C. Exploring the human microbiome from multiple perspectives: factors altering its composition and function. FEMS Microbiol Rev. 2017;41(4):453–78.
Walker AW, Duncan SH, Louis P, Flint HJ. Phylogeny, culturing, and metagenomics of the human gut microbiota. Trends Microbiol. 2014;22(5):267–74.
Bilen M, Dufour JC, Lagier JC, Cadoret F, Daoud Z, Dubourg G, Raoult D. The contribution of culturomics to the repertoire of isolated human bacterial and archaeal species. MICROBIOME. 2018;6(1):94.
Dubourg G, Baron S, Cadoret F, Couderc C, Fournier PE, Lagier JC, Raoult D. From Culturomics to clinical microbiology and forward. Emerg Infect Dis. 2018;24(9):1683–90.
Seck EH, Senghor B, Merhej V, Bachar D, Cadoret F, Robert C, Azhar EI, Yasir M, Bibi F, Jiman-Fatani AA, et al. Salt in stools is associated with obesity, gut halophilic microbiota and Akkermansia muciniphila depletion in humans. Int J Obes. 2019;43(4):862–71.
Gong S, Lan T, Zeng L, Luo H, Yang X, Li N, Chen X, Liu Z, Li R, Win S, et al. Gut microbiota mediates diurnal variation of acetaminophen induced acute liver injury in mice. J Hepatol. 2018;69(1):51–9.
Ghequire MG, De Mot R. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol Rev. 2014;38(4):523–68.
Jacobson AP, Wang H, Gill VS, Duvall R, Arce G, Chirtel S, Hammack TS. Relative effectiveness of selected preenrichment media for the detection of Salmonella from leafy green produce and herbs. Food Microbiol. 2017;63:123–8.
Wang H, Gill VS, Cheng CM, Gonzalez-Escalona N, Irvin KA, Zheng J, Bell RL, Jacobson AP, Hammack TS. Evaluation and comparison of rapid methods for the detection of Salmonella in naturally contaminated pine nuts using different pre enrichment media. Food Microbiol. 2015;46:58–65.
Cobian GA, Youle M, Cantu VA, Felts B, Nulton J, Rohwer F. Viruses as winners in the game of life. ANNU REV VIROL. 2016;3(1):197–214.
Tan JH, Vidaillac C, Yam J, Chua SL, Givskov M, Yang L: In Vitro and In Vivo Efficacy of an LpxC Inhibitor, CHIR-090, Alone or Combined with Colistin against Pseudomonas aeruginosa Biofilm. Antimicrob Agents Chemother 2017, 61(7).
Duncan SH, Hold GL, Harmsen HJ, Stewart CS, Flint HJ. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. Nov., comb. nov. Int J Syst Evol Microbiol. 2002;52(Pt 6):2141–6.
Konig J, Siebenhaar A, Hogenauer C, Arkkila P, Nieuwdorp M, Noren T, Ponsioen CY, Rosien U, Rossen NG, Satokari R, et al. Consensus report: faecal microbiota transfer - clinical applications and procedures. Aliment Pharmacol Ther. 2017;45(2):222–39.
Acknowledgments
We thank Louise Adam, ELS(D), from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac) for editing the English text of a draft of this manuscript.
Consent for publication
Not applicable.
Funding
This research is supported by National Natural Science Foundation for Key Programs of China Grants (No. 81790632), Science and Technology Program of Guangdong, China Grants (No.2017B030314037 & No.2017B020209003), Innovation Leader Team Program of Guangzhou (No. 201809010014) and Program on the innovation of environment construction of Guangzhou (No.201906010034). The funders had no role in design of study, data collection and analysis, and preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
FH did the experiments, analyzed the data, and wrote the manuscript; YC did the experiments and analyzed the data; ZH and NH analyzed data; LB and HD did the experiments and investigated the literature; ZL, ZP, LD and HG collected and pretreated samples; RY directed the experiments and contributed to revise the manuscript; FZ and YB designed the experiments, provided overall directions and contributed to revising the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The research was approved by the Ethics Committee of Shijitan Hospital (Beijing, China; agreement no. 2018KY55). And all volunteers signed a consent document and consented to isolate gut bacteria from their feces. All procedures were performed according to the guidelines of the Ethics Committee and in accordance with local laws and regulations.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1:
Supplementary table and figure.
Additional file 2: Table S2
Taxonomic information of 102 species of bacteria.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Hou, F., Chang, Y., Huang, Z. et al. Application of LpxC enzyme inhibitor to inhibit some fast-growing bacteria in human gut bacterial culturomics. BMC Microbiol 19, 308 (2019). https://doi.org/10.1186/s12866-019-1681-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-019-1681-6