Abstract
Background
The ability to eliminate a parental genome from a eukaryotic germ cell is a phenomenon observed mostly in hybrid organisms displaying an alternative propagation to sexual reproduction. For most taxa, the underlying cellular pathways and timing of the elimination process is only poorly understood. In the water frog hybrid Pelophylax esculentus (parental taxa are P. ridibundus and P. lessonae) the only described mechanism assumes that one parental genome is excluded from the germline during metamorphosis and prior to meiosis, while only second genome enters meiosis after endoreduplication. Our study of hybrids from a P. ridibundus—P. esculentus-male populations known for its production of more types of gametes shows that hybridogenetic mechanism of genome elimination is not uniform.
Results
Using comparative genomic hybridization (CGH) on mitotic and meiotic cell stages, we identified at least two pathways of meiotic mechanisms. One type of Pelophylax esculentus males provides supporting evidence of a premeiotic elimination of one parental genome. In several other males we record the presence of both parental genomes in the late phases of meiotic prophase I (diplotene) and metaphase I.
Conclusion
Some P. esculentus males have no genome elimination from the germ line prior to meiosis. Considering previous cytological and experimental evidence for a formation of both ridibundus and lessonae sperm within a single P. esculentus individual, we propose a hypothesis that genome elimination from the germline can either be postponed to the meiotic stages or absent altogether in these hybrids.
Similar content being viewed by others
Background
Meiosis is a vital process in all sexual organisms, ensuring fertility and genome stability and encouraging genetic diversity [14, 22]. Sexual reproduction involves the recombination of parental genomes followed by the coordinated segregation of the recombined chromosomes into gametes [57]. Despite the conservative nature of meiotic machinery, a number of anticipated mechanisms, including hybridization, can disrupt the regular cycles and alter the normal course of meiosis [41]. In hybrid animals, these deviations have resulted in a loss of sexual reproduction accompanied by modifications in gametogenesis such as premeiotic endomitosis (duplication of chromosomes), and genome exclusion (the loss of one parental genome) (reviewed in [26, 43]).
Hybridogenesis is a mode of bisexual reproduction characterized by the exclusion of one complete parental genome from the germline, while the remaining genome is endoreduplicated and subsequently transferred clonally (referred to as a hemiclone; [39, 55]). Hybridogenetic animals usually mate with the sexual species that contribute the eliminated genome [6, 9, 39]. New hybrids are generated via true fertilization, however, the genome from the sexual mate is discarded again in the next round of gamete formation.
Hybridogenesis has been recorded in the diploid all-female fish of the genus Poeciliopsis [39, 40], and Cimino [7, 8] observed the exclusion of P. lucida chromosomes during the onset of meiosis, while in P. monacha the genome is transferred into a reconstituted nucleus by the unipolar spindle. Apart from these species, very little is known about the cytological processes in other hybridogenetic or hybridogenesis-related animals such as the Squalius alburnoides fish [1], the Misgurnus anguillicaudatus fish [27], the Asian loach fish of the genus Cobitis [23], the carp gudgeon Hypseleotris [38], Ambystoma salamanders, Bufotes baturae toads [44], and Pelophylax esculentus water frogs [10, 17, 49].
The European sexual species Pelophylax lessonae and P. ridibundus hybridize and produce the hybrid form P. esculentus, which maintains a permanent F1 (first filial) hybrid state from generation to generation. This hybrid is able to exclude one parental genome from its germline and to duplicate the remaining one. As a result, the hybrid produces unrecombined ridibundus or lessonae gametes and therefore continues with only one parental species, i.e. the species whose genome has been eliminated (e.g. [2, 18, 47]).
It is generally believed that the exclusion of a parental genome from P. esculentus germ cells takes place before the onset of meiotic prophase I, followed by the endoreduplication of the remaining ridibundus genome [10, 11, 48]. In females the majority of oogonia have already been transformed into oocytes with 13 diplotene bivalents, usually by the time P. esculentus have entered their first hibernation [48]. Similarly, the proliferating spermatozoa in the testes of adult P. esculentus contained a diploid set of only ridibundus chromosomes [20]. Hence, the process of genome elimination and reduplication seems to occur at an early stage of spermatogenesis [20]. Further evidence comes from Günther [17], who observed in P. esculentus males from Eastern Germany a large number of meiotic figures with irregularities such as aneuploidy, univalency and heterologous multivalency. He interpreted his results as evidence contradicting the occurrence of a single cytological mechanism of hybridogenesis. Detailed cytological studies of male meiosis have yet to be carried out.
P. esculentus typically forms two reproductive systems; one with P. lessonae and one with P. ridibundus. The latter mostly consists of P. ridibundus (females and males) and only diploid hybrid males [50, 51]. Such P. ridibundus—P. esculentus-male populations have been found in Central Europe, mostly along the Oder River (reviewed by [34]). Here, hybrid males inherit either the lessonae or the ridibundus genome, or produce a combination of both kinds of sperm [3, 19, 35, 51, 54].
In order to understand the cytogenetic basis of these inheritance patterns, we studied the mitotic and meiotic cell stages of hybrids of a P. ridibundus—P. esculentus-male population from the Upper Oder River. Using comparative genomic hybridization (CGH) we discovered that the elimination of one parental genome does not necessarily precede meiotic divisions. In fact, the opposite is often true, where maintaining both parental genomes later in meiotic phases is actually relatively common.
Methods
Animals
We examined 14 adult and 4 subadult male individuals of P. esculentus from three different P. ridibundus—P. esculentus male populations along the Upper Oder River (49.914498, 18.091502; 49.705486, 18.092624; 49.735014, 18.152479). For genomic probes, we used two adult P. lessonae males (50.043063, 13.441079; 49.761259, 18.597399) and two adult P. ridibundus males from surrounding localities (49.705293, 18.081609). Specimens were genotyped using three polymorphic allozyme loci: Aspartate aminotransferase (Aat; EC 2.6.1.1), Glucose-6-phosphate isomerase (Gpi; EC 5.3.1.9) and Lactate dehydrogenase (Ldh-1; EC 1.1.1.27) [50]. All experimental procedures were conducted with the approval, and under the supervision of the Ethical Committee of the Faculty of Science,
Charles University, Prague, according to the directives of the State Veterinary Administration of the Czech Republic, permit number 34711/2010-30 from the Ministry of Agriculture of the Czech Republic. Specimens were deposited in the frog collection of the Laboratory of Fish Genetics, IAPG CAS, Liběchov. Permissions 358/2011 required for the field work collection of the frogs were obtained from the Agency for Nature Conservation and Landscape Protection of the Czech Republic.
Chromosome preparations
We employed two different protocols to obtain chromosome spreads from gonadal tissues. In the majority of adult and subadult individuals we adapted the protocol of Zaleśna et al. [56], originally designed for chromosome preparation from bone marrow. In juvenile specimens with small gonads we applied a spreading technique previously used for spiders [25] with slight modifications. Briefly: after the dissection of a juvenile specimen the gonads were removed and hypotonized in 0.075 M KCl for 8 min, followed by three rounds (15, 30, 60 min) of fixation in 3:1 methanol / acetic acid solution. The fixed gonadal tissue was then suspended in 60 % acetic acid and spread on a hot-plate (40 °C).
For conventional cytogenetic analysis, chromosomes were stained with 5 % Giemsa solution (pH 6.8) (Merck, Darmstadt, Germany). Selected slides were destained in methanol / acetic acid fixative, dehydrated in an ethanol series (70, 80, and 96 %, 3 min each) and stored in a freezer (-20 °C) for subsequent cytogenetic experiments.
DNA extraction and probe preparation
Whole genomic DNAs (gDNAs) from P. ridibundus and P. lessonae were extracted from muscle tissue using the conventional phenol-chloroform-isoamylalcohol method [13]. Probes prepared from both parental species were differentially labelled either with biotin-16-dUTP (2’-Deoxyuridine, 5’-Triphosphate, Roche, Mannheim, Germany) or digoxigenin-11-dUTP (Roche) using Nick Translation Mix (Abbott Molecular, Illinois, USA or Roche Diagnostics, Mannheim, Germany). For each slide, 1 μg of P. ridibundus gDNA, 1 μg of P. lessonae gDNA and 50 μg of sonicated salmon sperm DNA (Sigma-Aldrich) were added and the resulting probe was precipitated in 96 % ethanol, washed in 70 % ethanol, air-dried and re-dissolved in 25 μl of hybridization buffer (50 % formamide, 10 % dextran sulphate, 2× SSC (Standard saline buffer), 0.04 M NaPO4 (Sodium Phosphate) buffer, 0.1 % SDS, Denhardt’s reagent, see [29]). In some experiments, the final probe also included 15–30 μg of unlabelled species-specific competitive DNA prepared from P. esculentus gDNA using a Illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare, Buckinghamshire, UK), followed by sonication of the amplified product (40 cycles, 10 pulses, 100 % power) to approximate fragment size of 100–200 bp using the ultrasonic homogenizer Sonopuls HD 2070 (Bandelin Electric, Berlin, Germany).
Comparative genomic hybridization (CGH)
In order to identify the chromosome sets of particular parental species within a hybrid genome throughout the meiotic phases we performed the CGH method according to Bi and Bogart [4] with several modifications. After thermal aging (3–4 h at 37 °C and 1 h at 60 °C) the chromosomes were treated with RNase A (Sigma-Aldrich) (200 μg/ml in 2× SSC, 90 min, 37 °C) and then pepsin (50 μg/ml in 10 mM HCl, 3 min, 37 °C). The slides were denatured in 75 % formamide (pH 7.0) (Sigma-Aldrich) in 2× SSC at 74 °C for 3 min, and then immediately cooled and dehydrated in 70 % (cold), 80 % and 96 % (RT) ethanol. The hybridization mixture was denatured at 86 °C for 6 min. Hybridization was performed at 37 °C for 48–72 h. Post-hybridization washes were applied twice in 50 % formamide in 2× SSC (pH 7.0) at 42 °C for 5 min and three times in 1× SSC at 42 °C (7 min each). In order to block non-specific binding sites for streptavidin and anti-digoxigenin, the slides were incubated with 500 μl of 3 % BSA (Vector Labs, Burlington, Canada) in 4× SSC in 0.01 % Tween 20 at 37 °C for 20 min. The hybridization signal was detected using Anti-Digoxigenin-Rhodamine (Roche) and Streptavidin-FITC (fluorescein isothiocyanate; Invitrogen Life Technologies, San Diego, CA, USA) or alternatively with Anti-Digoxigenin-Fluorescein (Roche) and Streptavidin-Cy3 (Invitrogen Life Technologies), to exclude any influence of antibodies and/or fluorochromes. The slides were incubated with antibodies at 37 °C for 60 min in a dark humid chamber. Finally, the slides were washed four times (7 min each) in 4× SSC in 0.01 % Tween (pH 7.0) at 42 °C and mounted in antifade containing 1.5 μg/ml DAPI (4’, 6-diamidino-2-phenylindole; Cambio, Cambridge, United Kingdom).
Image processing
Chromosomal preparations were inspected using a Provis AX70 (Olympus) fluorescence microscope equipped with standard fluorescence filter sets. Selected images for each fluorescent dye were captured separately with a black and white CCD camera (DP30BW Olympus) using Olympus Acquisition Software. The digital images were then pseudocoloured (blue for DAPI, red for Rhodamine or Cy3, green for FITC) and superimposed using MicroImage software (Olympus, version 4.0). The images were optimized for brightness and contrast using Adobe Photoshop, version CS5.
Results
We obtained chromosomal preparations from the gonads of 18 male individuals. The preparations contained different phases of meiotic division as well as spermatogonial mitotic metaphases. Giemsa-stained karyotypes (not shown) confirmed the previous description of Zaleśna et al. [56], with all species of the Pelophylax hybridogenetic complex having 26 metacentric and submetacentric chromosomes. Moreover, in line with the findings from the mentioned study, the homologous chromosomes in P. esculentus differed slightly in size. Along with spermatogonial metaphases, we also observed stages with haploid or diploid chromosome numbers corresponding to particular meiotic and/or pre-meiotic phases (Fig. 1a-e). Haploid chromosome complements appeared to correspond to either a premeiotic stage after the elimination of one parental genome (Fig. 1b) or to chromosomes in the first meiotic division (Fig. 1d). Diploid chromosome complements represented either mitotic metaphases (Fig. 1a) or stages of the first meiotic division with bivalents (Fig. 1c).
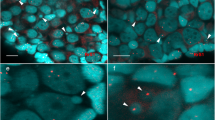
Comparative genomic hybridization (CGH) in mitotic and meiotic chromosomes of four water frog Pelophylax esculentus males. M1 (a), M2 (b-d), M3 (e-g, j) and M4 (h, i). CGH clearly distinguished chromosomes of the parental species, P. ridibundus (red) and P. lessonae (green). a Mitotic prometaphase. b Haploid mitotic metaphase after elimination of the ridibundus genome. c Diplotene. d Meiotic metaphase I. e, f, g, h Late meiotic prophase I. i, j Meiotic metaphase I showing bivalent-like configurations and univalents. Solid arrowheads indicate the smallest submetacentric chromosome pair with marked ridibundus-specific repetitive DNA in the lessonae-derived chromosome set, arrows indicate bivalent-like configurations between two different parental genomes, open arrowheads indicate bivalent-like configurations within one parental genome, asterisks indicate univalents. Scale bars equal 10 μm
We examined the mitotic and meiotic spreads further by means of CGH in four hybrid males (M1-M4). Although chromosome spreads were successfully obtained from all individuals, the hybridization procedure was only successful in four of them. Some examples of unsuccessful hybridization patterns are shown in Additional file 1: Figure S1-S3. A possible explanation for the general failure of CGH could be its high sensitivity in respect to experimental conditions [45, 46]. Multiple successful repetitions of the CGH experiments did however confirm that the chromosomal patterns observed in germinal cells of four esculentus males (M1-M4) were not artefacts. CGH provided a clear discrimination between the chromosomes of P. lessonae and P. ridibundus (Fig. 1a). The observed differential hybridization pattern of chromosome complements containing both parental genomes most probably resulted from the presence of species-specific repetitive sequences [24], very likely including some sort of transponable elements (TEs) and microsatellites [33]. Both experimental approaches (either with- or without the specific competitive DNA prepared from P. esculentus) yielded the same resulting hybridization pattern (Fig. 2a, b).
Mitotic metaphases of a Pelophylax esculentus male after comparative genomic hybridization (CGH). a CGH with specific competitive DNA prepared from P. esculentus. b CGH without specific competitive DNA. P. ridibundus chromosomes are visible as red signals, P. lessonae chromosomes as green signals. Scale bars equal 10 μm
Two groups of males were distinguishable by their differences in hybridization patterns. In the first group (male M2), nearly all chromosomes, with the exception of the smallest submetacentrics, were predominately highlighted with the lessonae-derived probe (Fig. 1b-d). The smallest submetacentric chromosome pair displayed a marked ridibundus-specific repetitive DNA region, even in the homologous lessonae-specific chromosomes (Fig. 1b, c, solid arrowheads). The number and morphology of the chromosomes indicated the presence of both mitotic (Fig. 1b) and meiotic stages (Fig. 1c, d). In the second group, 89 out of 122 chromosome complements (49 out of 55 in male M1, 18/22 in male M3, and 22/45 in male M4) showed a mixture of chromosomes with two different hybridization patterns, i.e. with strong hybridization signals of the lessonae-derived probe and the ridibundus-derived probe (Fig. 1a, e-j). All chromosomal complements showing both parental genomes were classified as diploid sets, either composed of mitotic chromosomes (Fig. 1a) or meiotic chromosomes in a late meiotic prophase I (Fig. 1e, f, g, h) or in a metaphase I (Fig. 1i, j).
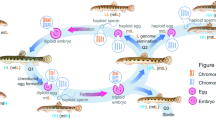
Based on the accurate identification of meiotic stages and on the scheme of hybridogenesis (Fig. 3) we tried to provisionally reconstruct the process of hybrid spermatogenesis. From 170 observed figures we identified five different mitotic or meiotic stages i.e. (i) mitotic metaphase with either diploid (Fig. 1a) or haploid (Fig. 1b) chromosome numbers, (ii) meiotic diplotene with regular bivalents (Fig. 1c) and (iii) meiotic metaphase MI (Fig. 1d) where 1c and 1d are composed of only one parental genome, (iv) late meiotic prophase I (Fig. 1e, f, g, h) and (v) meiotic metaphase MI (Fig. 1i, j) where chromosomes of both parental species formed bivalent-like configurations. More specifically, while male M2 exhibited only the lessonae-derived chromosomes in meiotic prophase I and metaphase I with 13 bivalents (each of them presumably composed of a pair of endoreduplicated identical chromosomes), the males M3 and M4 displayed chromosomes apparently derived from both parental genomes in their meiotic prophase I. These males formed bivalent-like configurations from non-homologous chromosomes that paired randomly either within (Fig. 1e, g, i, j, open arrowheads) or between parental genomes (Fig. 1e, g, i, j, arrows). Moreover, some chromosomes did not form a bivalent-like configuration, but instead remained unpaired as univalents (Fig. 1e, g, i, j, asterisks).
Schema of hybridogenesis assumed for maintenance of diploid hybrid male M2 (this study) in mixed populations with P. ridibundus. a elimination of the P. ridibundus genome (red); b reduplication of the P. lessonae genome (green). As a result haploid P. lessonae gametes are produced. The vertical solid arrow shows spermatogonia, the dashed arrow spermatocytes. Meiotic cycle starts after b
Discussion
Our analysis of the meiotic mechanism of Pelophylax esculentus males provides supporting evidence of premeiotic genome elimination. In addition to this observation, we record the presence of both parental genomes in the late phases of meiotic prophase I (diplotene) and metaphase I in several other males. Our results suggest that some males have no genome elimination from the germ line prior to meiosis.
The formation of clonal gametes during hybridogenetic spermatogenesis depends on a range of coordinated molecular and cytogenetic processes that are not yet fully understood. It is generally believed that in the germ cells of diploid hybrids one parental chromosome set is eliminated before entering the meiotic cycle, while the remaining set is endoreduplicated (e.g., [20]). This pattern was observed in at least one hybrid male (M2; Fig. 1b-d). The meiotic divisions obtained from this male contained only green coloured lessonae chromosomes either in a haploid set, after the elimination of the red coloured ridibundus chromosomes, Fig. 1b), or in a diploid number, after genome duplication (Fig. 1c-d). Such an inheritance mode would lead to sperm with a lessonae genome, which would mean that after fertilization of the P. ridibundus egg the F1 hybrid state would be restored. As the meiotic chromosomes treated with comparative genomic hybridization (CGH) did not display any recombination between the lessonae and ridibundus chromosomes such as crossing-over or other types of recombination, this male must have transferred its lessonae genome clonally into its sperm as assumed for hybrid males from P. ridibundus—P. esculentus-male populations [19, 51].
A completely different pattern of spermatogenesis was found in males M3 and M4 where the majority of nuclei in the first meiotic division contained both ridibundus and lessonae chromosome sets. Most of the nuclei were in the late meiotic prophase I, probably corresponding to diplotene (Fig. 1e, f, g, h) with some of them even reaching metaphase I (Fig. 1i, j). This finding clearly suggests that the majority of spermatocytes did not carry out genome elimination prior to meiosis. Previous studies based on protein electrophoresis have indicated that in the germ line of P. esculentus genome elimination takes place before meiosis [12, 20, 52], likely during the last mitotic division [48] in the so called “E” (Elimination) phase [53]. There are two principle hypotheses concerning genome exclusion: 1) an exclusion takes place during the mitotic phase whereby the excluded genome is enzymatically degraded [31, 54], or 2) the elimination of whole chromosomes, or at least parts of them, takes place during mitosis of the gametogonia [31]. The latter hypothesis seems less likely as no irregularities in the spindle apparatus or in the heterochromatization have been observed (see pp. 91–92 of [34]). It is not yet clear whether genome elimination is a one-step or a gradual process during mitotic division [31]. Within vertebrates, only the all-female fish of the genus Poeciliopsis eliminate one chromosome set as late as in meiosis but even in this fish it occurs during prophase I [7, 8].
The occurrence of both parental genomes in the proliferating spermatozoa of P. esculentus investigated in this study conflicts with our expectation of observing only one parental genome in the meiotic cells of adult males [20]. It further suggests that the elimination phase (if present) is not restricted to the period around metamorphosis.
Using conventional cytogenetic techniques, the absence of genome exclusion has been assumed in some hybrids from P. ridibundus—P. esculentus-male populations [17, 21] and in just a single laboratory-synthesized P. esculentus male [36]. The related observations of numerous aberrations during meiosis in P. esculentus males such as aneuploidy, degenerated chromosomes and heterologous multivalents [17, 32] and of fertility disorders in many P. esculentus males (e.g. [15, 16, 30]) can be considered as evidence for selection processes acting during pregametic and/or gametic stages [19]. As well as cell lineages in which one parental genome is excluded premeiotically, lineages (spermatogonia, spermatocytes) with both parental genomes may undergo cellular selection during meiosis. As a result, lineages with balanced genomes (probably with the chromosomes of only one parental species) may yield fertile sperm while those with unbalanced haploid genomes (a mixture of lessonae and ridibundus chromosomes) would result in infertile sperm [19].
Indeed, irregular diplotene stages (Fig. 1e, g, i, j) with bivalent-like configurations and univalents, and the fact that most ridibundus chromosomes paired with non-homologous ridibundus chromosomes rather than with homologous lessonae chromosomes and vice-versa, may indicate malfunctions in the process of genome haploidization and meiosis in general. But in terms of the number of chromosomes, meiotic prophase I with 13 ridibundus and 13 lessonae chromosomes (Fig. 1i, j) did not differ from regular meiotic phases with 13 bivalents. More thorough analyses are necessary to understand whether such cells may or not produce functional sperm. Currently, two alternative hypotheses remain open. First, such cells may still result in dysfunctional sperms [19]. It was already observed that many P. esculentus males exhibit degenerated testes, low numbers of sperm, high numbers of immobilized and/or inhibited sperm [19, 30, 37]. Second, the cells may yield both unrecombined lessonae and ridibundus sperm [19, 51, 54]. Vinogradov et al. [54] recorded “so-called hybrid amphispermy” in 14–17 % of P. esculentus males. Although the underlying cytogenetic mechanisms were not identified, in principle, two mechanisms are conceivable: 1) genome exclusion is unspecific and takes place during meiosis leading to clonal cell lineages with only lessonae or ridibundus chromosomes, or 2) the chromosomes are segregated non-randomly during meiosis, probably in anaphase I, i.e. without interchromosomal recombination, resulting in both lessonae and ridibundus spermatids and sperms.
Chromosomal studies of deviations from canonical gametogenesis in P. esculentus females have shown observations of very rare oocytes in which elimination has not occurred [5, 10] resembling the mechanism of premeiotic endoreplication in automictic parthenogenesis [28, 42]. Dedukh et al. [10] also observed aneuploid oocytes suggesting a partial loss of chromosomes during gametogenesis. Together with our observations that some diploid P. esculentus males have no genome elimination from the germ line prior to meiosis, the phenomenon of no chromosome elimination may be more common than previously thought.
Conclusions
The central finding of this study is that genome elimination in P. esculentus males is not always restricted to larval or juvenile stages, as both parental genomes were discovered to still be present in the germline of the adult specimens. We propose the following three hypotheses about the fate of homologous and non-homologous bivalent-like configurations of lessonae and ridibundus chromosomes observed in the first meiotic division: 1) such bivalents represent a process leading to unviable gametes; 2) the elimination phase is postponed to later stages of the meiotic cell cycle; 3) there is no genome elimination, homologous lessonae and ridibundus chromosomes segregate in anaphase I resulting in both haploid lessonae and ridibundus sperm.
Overall, our data provide new information about the behavior of two species-specific genomes in the meiotic cycle which will help us understand the underlying cytogenetic mechanisms regulating the formation of clonal gametes. As the molecular mechanisms leading to genome exclusion and subsequent gamete formation are still unclear, not only in water frogs but also in other asexuals, further research should focus on the mechanisms of homologous chromosome pairing and segregation in later meiotic phases.
Abbreviations
Aat, aspartate aminotransferase; Cy3, cyanine dye; CGH, comparative genomic hybridization; DAPI, 4’, 6-diamidino-2-phenylindole; dUTP, 2’-Deoxyuridine, 5’-Triphosphate; E, elimination; F1, first filial generation; FITC, fluorescein isothiocyanate; gDNA, whole genomic DNA; Gpi, Glucose-6-phosphate isomerase; HCl, hydrogen chloride; IAPG CAS, v.v.i., Institute of Animal Physiology and Genetics of the Czech Academy of Sciences, v.v.i.; KCl, kalcium chloride; Ldh-1, lactate dehydrogenase; NaPO4, sodium phosphate; SDS, sodium dodecyl sulfate used as Denhardt’s reagent; SSC, Standard saline buffer; TEs, transposable elements
References
Alves MJ, Coelho MM, Collares-Pereira MJ. Evolution in action through hybridisation and polyploidy in an Iberian freshwater fish: a genetic review. Genetica. 2001;111:375–85.
Berger L, Günther R. Genetic composition and reproduction of water frog populations (Rana kl. esculenta Synklepton) near nature reserve Serrahn, GDR. Arch Natschutz Landschforsch Berlin. 1988;28:265–80.
Berger L, Günther R. Inheritance patterns of water frog males from the environments of nature reserve Steckby, Germany. Zool Pol. 1991–1992;37:87–100.
Bi K, Bogart JP. Identification of intergenomic recombination in unisexual salamanders of the genus Ambystoma by genomic in situ hybridization (GISH). Cytogenet Genome Res. 2006;112:307–12.
Bucci S, Ragghianti M, Mancino G, Berger L, Hotz H, Uzzell T. Lampbrush and mitotic chromosomes of the hemiclonally reproducing hybrid Rana esculenta and its parental species. J Exp Zool. 1990;255:37–56.
Choleva L, Janko K, De Gelas K, Bohlen J, Šlechtová V, Rábová M, Ráb P. Synthesis of clonality and polyploidy in vertebrate animals by hybridization between two sexual species. Evolution. 2012;66:2191–203.
Cimino MC. Egg-production, polyploidization and evolution in a diploid all–female fish of the genus Poeciliopsis. Evolution. 1972a;26:294–306.
Cimino MC. Meiosis in triploid all–female fish (Poeciliopsis, Poeciliidae). Science. 1972b;175:1484–1486.
Dawley RM. An introduction to unisexual vertebrates. In: Dawley RM, Bogard JP, editors. Evolution and Ecology of unisexual vertebrates New York State museum, vol. 466. Albany: New York Bulletin; 1989. p. 1–18.
Dedukh D, Litvinchuk SN, Rosanov JM, Shabanov DA, Krasikova AK. Crossing experiments reveal gamete contribution into appearance of di–and triploid hybrid frogs in Pelophylax esculentus population systems. Chromosome Res. 2015;23:380–1.
Graf JD, Müller WP. Experimental gynogenesis provides evidence of hybridogenetic reproduction in the Rana esculenta complex. Experientia. 1979;35:1574–6.
Graf JD, Karch F, Moreillon MC. Biochemical variation in the Rana esculenta complex: A new hybrid form related to Rana perezi and Rana ridibunda. Experientia. 1977;33:1582–4.
Graham DE. The isolation of high molecular weight DNA from whole organisms or large tissue masses. Anal Biochem. 1978;85:609–13.
Grandont L, Jenczewski E, Lloyd A. Meiosis and its deviations in polyploid plants. Cytogenet Genome Res. 2013;140:171–84.
Günther R. Der Karyotyp von Rana ridibunda Pall. und das Vorkommen von Triploidie bei Rana esculenta L. (Anura, Amphibia). Biol Zentralbl. 1970;89:327–42.
Günther R. Über die verwandtschaftlichen Beziehungen zwischen den europäischen Grünfröschen und den Bastardcharakter von Rana esculenta L. (Anura). Zool Anz. 1973;190:250–85.
Günther R. Untersuchungen der Meiose bei Männchen von Rana ridibunda Pall., Rana lessonae Cam. und der Bastardform “Rana esculenta” L. (Anura). Biol Zentralbl. 1975;94:277–94.
Günther R. Zur Populationsgenetik der mitteleuropäischen Wasserfröschen des Rana esculenta–Synkleptons (Anura, Ranidae). Zool Anz. 1983;197:43–54.
Günther R, Plötner J. Zur Problematik der klonalen Vererbung bei Rana kl. esculenta (Anura). In: Beiträge zur Biologie und Bibliographie (1960–1987) der europäischen Wasserfrösche. Jb Feldherp Beiheft. 1988;1:23–46.
Heppich S, Tunner HG, Greilhuber J. Premeiotic chromosome doubling after genome elimination during spermatogenesis of the species hybrid Rana esculenta. Theor Appl Genet. 1982;61:101–4.
Hotz H, Uzzell T. Interspecific hybrids of Rana ridibunda without germ line exclusion of a parental genome. Experientia. 1983;39:538–40.
John B. Meiosis. 3rd ed. Cambridge: Cambridge University Press; 1990.
Kim IS, Lee EH. Hybridization experiment of diploid–triploid cobitid fishes, Cobitis sinensis-longicorpus complex (Pisces: Cobitidae). Folia Zool. 2000;49:17–22.
Kato A, Vega JM, Han F, Lamb JC, Birchler JA. Advances in plant chromosome identification and cytogenetic techniques. Curr Opin Plant Biol. 2005;8:148–54.
Král J, Musilová J, Št’áhlavský F, Řezáč M, Akan Z, Edwards RL, et al. Evolution of the karyotype and sex chromosome systems in basal clades of araneomorph spiders (Araneae: Araneomorphae). Chromosome Res. 2006;14:859–80.
Lamatsch DK, Stöck M. Lost sex. Netherlands: Springer; 2009.
Morishima K, Yoshikawa H, Arai K. Meiotic hybridogenesis in triploid Misgurnus loach derived from a clonal lineage. Heredity. 2008;100:581–6.
Neaves WB, Baumann P. Unisexual reproduction among vertebrates. Trends Genet. 2011;27:81–8.
Neusser M. Karyotypevolution, Genomorganisation und Zellkernarchitektur der Neuweltaffen (Doctoral dissertation, lmu). 2004.
Ogielska M, Bartmańska J. Development of testes and differentiation of germ cells in water frogs of the Rana esculenta-complex (Amphibia, Anura). Amphibia Reptilia. 1999;20:251–63.
Ogielska M. Nucleus–like bodies in gonial cells of Rana esculenta [Amphibia, Anura] tadpoles-a putative way of chromosome elimination. Zool Pol. 1994;39:461–74.
Ohtani H. Mechanism of chromosome elimination in the hybridogenetic spermatogenesis of allotriploid males between Japanese and European water frogs. Chromosoma. 1993;102:158–62.
Plötner J, Köhler F, Uzzell T, Beerli P, Schreiber R, Guex GD, Hotz H. Evolution of serum albumin intron-1 is shaped by a 5’ truncated non–long terminal repeat retrotransposon in western Palearctic water frogs (Neobatrachia). Mol Phylogenet Evol. 2009;53:784–91.
Plötner J. Die westpaläarktischen Wasserfrösche: von Märtyrern der Wissenschaft zur biologischen Sensation. Germany: Laurenti; 2005.
Polls Pelaz M. Modes of gametogenesis among kleptons of the hybridogenetic water frog complex: an evolutionary synthesis. Zool Pol. 1994;39:123–38.
Ragghianti M, Bucci S, Marracci S, Casola C, Mancino G, Hotz H, et al. Gametogenesis of intergroup hybrids of hemiclonal frogs. Genet Res. 2007;89:39–45.
Reyer HU, Niederer B, Hettyey A. Variation in fertilisation abilities between hemiclonal hybrid and sexual parental males of sympatric water frogs (Rana lessonae, R. esculenta, R. ridibunda). Behav Ecol Sociobiol. 2003;54:274–84.
Schmidt DJ, Bond NR, Adams M, Hughes JM. Cytonuclear evidence for hybridogenetic reproduction in natural populations of the Australian carp gudgeon (Hypseleotris: Eleotridae). Mol Ecol. 2011;20:3367–80.
Schultz RJ. Hybridization, unisexuality, polyploidy in the teleost Poeciliopsis (Poeciliidae) and other vertebrates. Am Nat. 1969;103(934):605–19.
Schultz R. Evolution ecology of unisexual fishes. Evol Biol. 1977;10:277–331.
Schurko AM, Neiman M, Logsdon JM. Signs of sex: what we know and how we know it. Trends Ecol Evolut. 2009;24:208–17.
Stenberg P, Saura A. Cytology of asexual animals. In: Lost Sex. Netherlands: Springer; 2009. p. 63–74.
Stenberg P, Saura A. Meiosis and its deviations in polyploid animals. Cytogenet Genome Res. 2013;140:185–203.
Stöck M, Ustinova J, Betto-Colliard C, Schartl M, Moritz C, Perrin N. Simultaneous Mendelian and clonal genome transmission in a sexually reproducing, all–triploid vertebrate. Proc R Soc Lond B Biol Sci. 2011;279:1293–9.
Symonová R, Sember A, Majtánová Z, Ráb P. Characterization of Fish Genomes by GISH and CGH. In: Fish Cytogenetic Techniques: Ray-Fin Fishes and Chondrichthyans. USA: CRC Press; 2015. p. 118.
Traut W, Winking H. Meiotic chromosomes and stages of sex chromosome evolution in fish: zebrafish, platyfish and guppy. Chromosome Res. 2001;9:659–72.
Tunner H. Die klonale Struktur einer Wasserfröschpopulation. Z Zool Syst Evolut forsch. 1974;12:309–14.
Tunner H, Heppich S. Premeiotic genome exclusion during oogenesis in the common edible frog, Rana esculenta. Naturwissenschaften. 1981;68:207–8.
Tunner H, Heppich-Tunner S. Genom exclusion and two strategies of chromosome duplication in oogenesis of a hybrid frog. Naturwissenschaften. 1991;78:32–4.
Uzzell T, Berger L. Electrophoretic phenotypes of Rana ridibunda, Rana lessonae, and their hybridogenetic associate, Rana esculenta. Proc Acad Nat Sci Phila. 1975;127:13–24.
Uzzell T, Günther R, Berger L. Rana ridibunda and Rana esculenta: a leaky hybridogenetic system (Amphibia Salientia). Proc Acad Nat Sci Phila. 1977;128:147–171.
Uzzell T, Hotz H, Berger L. Genome exclusion in gametogenesis by an interspecific Rana hybrid: evidence from electrophoresis of individual oocytes. J Exp Zool. 1980;214:251–9.
Vinogradov AE, Borkin LJ, Günther R, Rosanov JM. Genome elimination in diploid and triploid Rana esculenta males: cytological evidence from DNA flow cytometry. Genome. 1990;33:619–27.
Vinogradov AE, Borkin LJ, Günther R, Rosanov JM. Two germ cell lineages with genomes of different species in one and the same animal. Hereditas. 1991;114:245–51.
Vrijenhoek RC, Angus RA, Schultz RJ. Variation heterozygosity in sexually vs. clonally reproducing populations of Poeciliopsis. Evolution. 1977;31:767–81.
Zaleśna A, Choleva L, Ogielska M, Rábová M, Marec F, Ráb P. Evidence for integrity of parental genomes in the diploid hybridogenetic water frog Pelophylax esculentus by genomic in situ hybridization. Cytogenet Genome Res. 2011;134:206–12.
Zielinski ML, Scheid OM. Meiosis in polyploid plants. In: Polyploidy and genome evolution. Berlin Heidelberg: Springer; 2012. p. 33–55.
Acknowledgements
We thank Mgr. Marie Altmanová for help with processing images using Adobe Photoshop. We thank Chris Johnson who proofread the manuscript.
Funding
MD and LC was supported by Grant No. 15-19947Y from The Czech Science Foundation and Grant No. 43-251468 from the Charles University Grant Agency . FM was supported by Grant No. 14-22765S from The Czech Science Foundation. MD, AS, PR and LC received institutional support No. RVO 67985904 from the Czech Academy of Sciences.
Availability of data and materials
All important data are provided in the Results and Figures. The dataset includes all the figures used to reach the conclusions drawn in the manuscript, and any additional data required to replicate the reported study findings in their entirety.
Authors’ contributions
MD participated in the design of the study, collected samples, made chromosomal preparations, participated in the in situ hybridization analysis and wrote the initial draft of the manuscript. AS performed the in situ hybridization analysis and drafted the manuscript. FM, PR and JP participated in the data interpretation and helped to draft the manuscript. LC conceived of the study, and participated in its design, sampling, and helped to draft the manuscript. All authors read and approved the final manuscript.
Competing interests
All authors declare no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All experimental procedures involving water frogs were performed in agreement with directives and under the supervision of the Ethical Committee of the Faculty of Science, Charles University, Prague, according to the directives of the State Veterinary Administration of the Czech Republic, permit number 34711/2010-30 from the Ministry of Agriculture of the Czech Republic. All institutional and national guidelines for the care and use of laboratory animals were followed.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1: Figure S1-S3.
Comparative genomic hybridization (CGH) on mitotic (1) and meiotic (2, 3) chromosomes of Pelophylax esculentus males showing several types of experimental artefacts and failures. 1) Unsuccessful differentiation of parental chromosomes: note the apparent accumulation of probes on the edges/surface of chromosomes, possibly due to over fixed gonadal tissues used for chromosome spreads. 2) Inconclusive hybridization pattern: note equal hybridization intensity of both genome-derived probes. 3) Week hybridization pattern, insufficient for differentiation of parental chromosomes. Lessonae-derived genomic probes were labelled with biotin-16-dUTP and hybridization signals detected with Streptavidin-FITC (green) (1a, 2a, 3a), ridibundus-derived genomic probes (b) with digoxigenin-11-dUTP and Anti-Digoxigenin-Rhodamine (red) (1b, 2b, 3b). Figures 1c, 2c, 3c show merged images of both genomic probes, figures 1d, 2d, 3d merged images of both probes and DAPI staining of chromosomes (blue). Scale bar = 10 μm. (TIF 2427 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Doležálková, M., Sember, A., Marec, F. et al. Is premeiotic genome elimination an exclusive mechanism for hemiclonal reproduction in hybrid males of the genus Pelophylax?. BMC Genet 17, 100 (2016). https://doi.org/10.1186/s12863-016-0408-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12863-016-0408-z