Abstract
Background
Glycosphingolipids (GSLs) are important membrane components composed of a carbohydrate structure attached to a hydrophobic ceramide. They can serve as specific membrane receptors for microbes and microbial products, such as F4 Escherichia coli (F4 ETEC) and isolated F4 fimbriae. The aim of this study was to investigate the hypothesis that variation in genes involved in the assembly of the F4 binding carbohydrate moiety of GSLs (i.e. ARSA, B4GALT6, GAL3ST1, GALC, GBA, GLA, GLB1, GLB1L, NEU1, NEU2, UGCG, UGT8) could account for differential binding of F4 ETEC and their fimbriae.
Results
RT-PCR could not reveal any differential expression of the 12 genes in the jejunum of F4 receptor-positive (F4R+) and F4 receptor-negative (F4R-) pigs. Sequencing the complete open reading frame of the 11 expressed genes (NEU2 was not expressed) identified 72 mutations. Although some of them might have a structural effect, none of them could be associated with a F4R phenotype.
Conclusion
We conclude that no regulatory or structural variation in any of the investigated genes is responsible for the genetic susceptibility of pigs towards F4 ETEC.
Similar content being viewed by others
Background
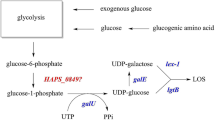
Glycosphingolipids (GSLs) are membrane components that participate in many intracellular and extracellular biological processes [1]. They are located in the outer leaflet of the plasma membrane in mammalian cells and are composed of a carbohydrate moiety linked to a lipid (ceramide). Biosynthesis of GSL occurs by the stepwise addition of carbohydrates first to the ceramide component, then to the growing carbohydrate chain [2]. The genes from the cerebroside-sulfatid region of the sphingolipid metabolism pathway are directly involved in synthesizing the carbohydrate core structure of GSLs (Figure 1).
F4 ETEC binding on GSLs and the 12 investigated genes of the cerebroside-sulfatid pathway. The carbohydrate moiety of GSLs has been shown to bind F4 ETEC and their fimbriae. According to Coddens et al. [4], galactosylceramide Galβ1Cer binds to F4ab/ac ETEC and fimbriae. Twelve genes involved in the carbohydrate moiety assembly of glycosphingolipids were selected from the cerebroside-sulfatid region of the sphingolipid metabolism pathway (adapted from KEGG pathway 00600). The solid lines represent molecular interaction or relation, the dashed lines represent linked to another map (see http://www.genome.jp/kegg-bin/show_pathway%3Fmap00600 for further details).
The cell surface carbohydrate structure of GSL can serve as specific binding sites for pathogens and their toxins, leading to subsequent adhesion [3]. Recently, it has been shown that the carbohydrate moiety of GSL interacts with F4 enterotoxigenic Escherichia coli (F4 ETEC) and their fimbriae [4]. F4 ETEC infections are a major cause of neonatal and post-weaning diarrhea in pigs [5]. Following attachment with their F4 fimbriae to specific receptors in the small intestine, they colonize the small intestine and produce enterotoxins (heat-labile and heat-stabile enterotoxins) which stimulate fluid secretion of epithelial cells, causing diarrhea in young pigs. Three antigenic F4 variants (F4ab, F4ac and F4ad) have been described [6]. The F4ac variant is worldwide the most common variant, except in central China where the F4ad variant is the most prevalent [5],[7]. Susceptibility towards F4 ETEC is inherited as an autosomal dominant Mendelian trait and the locus controlling F4ab/ac ETEC susceptibility has been mapped on chromosome 13. Recently, a new refined candidate region for F4ab/ac ETEC susceptibility has been identified on chromosome 13 indicating that the causal mutation for F4ab/ac ETEC susceptibility is not located in the previous suggested candidate genes on chromosome 13 [8]. The locus controlling F4ad ETEC susceptibility has not been mapped yet. So far, no causal mutation explaining the F4ab/ac/ad ETEC susceptibility in pigs has been identified [8]-[10]. Therefore, the purpose of this study is to determine if the F4 ETEC binding differences observed by Coddens et al. [4] could be explained by differential expression (for F4ab/ac/ad) or structural variation (for F4ad) of genes involved in the assembly of the carbohydrate moiety of GSLs (Figure 1).
Results and discussion
For all 12 genes (Additional file 1: Table S1) there is a curated human reference sequence available in the public databases. In pig however, this is so far only the case for GALC. For 9 genes (i.e. ARSA, B4GALT6, GAL3ST1, GBA, GLA, GLB1L, NEU1, NEU2 and UGCG) there was a predicted porcine sequence. These sequences were subjected to an in silico gene analysis and experimental validation. The coding sequence of all predicted porcine sequences was found to be correct since the exact sequence was found to be expressed in the jejunum, except for NEU2 that was not expressed. We neither observed NEU2 expression in porcine lymph node, heart, lung, dorsal muscle, diaphragm, liver, spleen, gall bladder, kidney, adrenal gland, bladder, duodenum, jejunum, ileum, colon and rectum (NEU2 assay was validated with DNA as a template; data not shown), which resembles the situation in human where extremely low levels of mRNA expression were found in all human tissues, except for testis, placenta and ovary [11]. Interspecies sequence comparison revealed the complete porcine GLB1 coding sequence in a non-annotated mRNA sequence [GenBank:AMP010068C04]. The exact sequence of 1992 bp (encoding a protein of 663 amino acids) was found to be expressed in the jejunum and shows 85% sequence identity with its human ortholog [GenBank: NM_000404.2]. The complete coding sequence of porcine UGT8 (1623 bp, encoding a protein of 541 amino acids) was amplified by RT-PCR from jejunum cDNA with primers based on its human ortholog [GenBank: NM_001128174.1]. Interspecies comparison showed only high sequence identities with UGT8 orthologs (93% with its human ortholog) and the sequence was submitted to NCBI as the first porcine UGT8 mRNA sequence [GenBank:JQ65026].
The eight pigs used in this study were solely phenotyped based on the in vitro villous adhesion test that has been proven to be reliable [12]-[14]. Phenotyping of the pigs based on the associated markers identified in previous linkage studies or based on the associated mutations in MUC4 and MUC13 would not be precise, because they are not in complete linkage disequilibrium with the F4ab/ac locus [8],[15]-[17]. Although linkage studies mapped the causal locus for the F4ab/ac susceptibility on chromosome 13 [8]-[10], it is possible that the expression of any of the 12 investigated genes is influenced by a trans-acting element present in this candidate region [8]. As no positional information is available for the F4ad ETEC receptor, a regulatory mutation impairing expression of any of the investigated GSL genes could also be responsible for the F4ad ETEC susceptibility in pigs. Because an obvious difference in expression between F4 receptor-positive (F4R+) and F4 receptor-negative (F4R-) pigs was expected, semi-quantitative measurements using 8 pigs with different F4 adhesion phenotypes were performed. For every amplicon a single fragment was generated with the same intensity for all samples. We can conclude that F4 ETEC susceptibility is not caused by any mutation affecting the expression level of any of the investigated genes nor by the expression of splice variants.
All amplicons generated in the expression study were sequenced to investigate if a structural mutation in any of these genes could be responsible for F4ad ETEC susceptibility. In total, 72 mutations were found: 45 silent mutations, 24 missense mutations, 2 mutations in the 3′UTR and 1 nonsense mutation (Additional file 2: Table S2). Only the silent mutation c.979 T > C in GALC was differential for the presence of the F4ad receptor in this sample set. The CC homozygotes and CT heterozygotes were present in the F4adR+ pigs and only TT homozygotes were present in the F4R- pigs. We expected a homozygous genotype in the F4adR- pigs, because resistance to F4 adhesion (F4R-) is inherited in a recessive Mendelian way [18]. We screened this mutation in 14 additional F4ad phenotyped pigs. Four TT homozygotes and 3 CT heterozygotes were observed in the F4adR+ pigs (n = 7) and 7 TT homozygotes in de F4adR- group (n = 7). Because 4 TT homozygotes were present in the F4adR+ pigs, we can conclude that this mutation is not associated with F4ad ETEC susceptibility. For completeness we also looked for association with the F4ab/acR phenotype, but as could be expected from the chromosomal position of the GSL genes none of the 72 mutations were differential in F4ab/acR+ and F4ab/acR- pigs.
Conclusions
Overall, we can conclude that no structural or regulatory variation in any of the 12 investigated genes is associated with F4 ETEC susceptibility. However, some of the mutations found (e.g. a nonsense mutation (c.1577C > G) in exon 5 of GLB1, introducing a premature stop codon (R656X) truncating the GLB1 protein with 8 amino acids at the C-terminus) may be of importance for other GSL-related diseases [19]-[21].
Methods
Sample collection
Crossbred pigs from different litters were euthanized at 5-18 weeks of age. Before euthanasia, blood samples were collected in EDTA blood tubes and stored at -20°C for DNA isolation. After slaughter, samples of mid-jejunum were collected using protocols approved by the animal care and ethics committee of the Faculty of Veterinary Medicine, University of Ghent (EC2010/042). Mid-jejunum samples for RNA isolation were washed three times with Krebs-Henseleit buffer (0.12 M NaCl, 0.014 M KCl, 0.001 M KH2PO4, 0.025 M NaHCO3, pH 7.4), immediately frozen in liquid nitrogen and stored at -80°C until RNA isolation. Villi from mid-jejunum samples for the in vitro villous adhesion assay were isolated and stored as described by Van den Broeck et al. [12].
Animal selection based on the in vitro villous adhesion assay
The in vitro villous adhesion assay for F4ab/ac/ad ETEC was carried out as described by Van den Broeck et al. [12]. Adhesion of more than 30 bacteria per 250 μm villous brush border length was noted as strong adhesive for F4 ETEC (F4R+) and less than 5 bacteria per 250 μm brush border length was noted as non-adhesive for F4 ETEC (F4R-) [14].
Eight pigs, representing 6 different F4 adhesion phenotypes, were selected for the expression study and mutation detection. These phenotypes were previously described as phenotype A (F4ab/ac/adR+; pig 1 and 2), B (F4ab/acR+; pig 3), C (F4ab/adR+; pig 4), D (F4adR+; pig 5), E (F4ab/ac/adR-; pig 6 and 7) and F (F4abR+; pig 8) [22],[23]. The phenotypes G (F4acR+) and H (F4ac/adR+), mainly observed in eastern breeds, were absent in our study [24]-[26]. Fourteen additional pigs were selected, only based on the presence of the F4ad receptor (7 F4adR+ and 7 F4adR-), for the GALC (c.979 T > C) mutation screening.
DNA isolation, RNA isolation and cDNA synthesis
DNA isolation from frozen blood samples was performed as described by Van Poucke et al. [27]. RNA isolation and cDNA synthesis of frozen mid-jejunum samples was performed as described by Goetstouwers et al. [28].
In silico gene analysis and experimental validation
Non-curated porcine gene sequences (Additional file 1: Table S1) from NCBI databases were (re)checked manually using BLAST analysis (genomic and mRNA) for a human-pig and a pig-pig comparison [29]. Primers were designed with Primer3Plus [30], generating overlapping amplicons that cover the complete coding sequence. RT-PCR products were generated with porcine mid-jejunum cDNA as a template (Additional file 3), of which 2 μl was used to check the amplicon length using agarose gel electrophoresis. The rest of the product (8 μl) was cleaned up with 4 U Exonuclease I and 2 U Antarctic Phosphatase (New England Biolabs) at 37°C for 30 min and 80°C for 15 min, and sequenced for verification. Forward and reverse sequencing reactions were performed with the PCR primers as described by Goetstouwers et al. [28].
Semi-quantitative expression study via RT-PCR
All above mentioned primers, generating overlapping amplicons covering the complete open reading frame of the 12 investigated genes, were used to perform RT-PCR (see above) on cDNA of the mid-jejunum samples of the 8 selected animals. Agarose gel electrophoresis was used to analyse the number and the length of the PCR products to check for phenotype explaining alternative splicing, and to compare the intensity of the bands to check for phenotype explaining differential expression (semi-quantitatively). ACTB was used as a validated reference gene [31].
Mutation detection via sequencing of the RT-PCR products
All RT-PCR products from the expression study were sequenced (see above) to check for F4ad phenotype explaining structural mutations.
GALC (c.979 T > C) mutation screening
The GALC (c.979 T > C) mutation was screened in 14 additional F4ad phenotyped pigs (7 F4adR+ and 7 F4adR-) via PCR with primer pair SscrGALC ± 4 and DNA as a template (Additional file 1: Table S1), and direct sequencing with the reverse sequence primer after PCR amplicon clean-up (see above).
Availability of supporting data
The data sets supporting the article are included within the article and its additional files.
Additional files
References
Lingwood CA: Glycosphingolipid functions. Cold Spring Harb Perspect Biol. 2011, 3: a004788-10.1101/cshperspect.a011874.
Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME: Essentials of Glycobiology. 2009, Cold Spring Harbor Press, New York (NY)
Schengrund CL: “Multivalent” saccharides: Development of new approaches for inhibiting the effects of glycosphingolipid-binding pathogens. Biochem Pharmacol. 2003, 65: 699-707. 10.1016/S0006-2952(02)01553-8.
Coddens A, Valis E, Benktander J, ngstr m J, Breimer ME, Cox E, Teneberg S: Erythrocyte and Porcine Intestinal Glycosphingolipids Recognized by F4 Fimbriae of Enterotoxigenic Escherichia coli. PLoS One. 2011, 6: 9-10.1371/journal.pone.0023309.
Fairbrother JM, Nadeau E, Gyles CL: Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim Health Res Rev. 2005, 6: 17-39. 10.1079/AHR2005105.
Gaastra W, de Graaf FK: Host-specific fimbrial adhesins of noninvasive enterotoxigenic Escherichia coli strains. Microbiol Rev. 1982, 46: 129-161.
Wang J, Jiang SW, Chen XH, Liu ZL, Peng J: Prevalence of fimbrial antigen (K88 variants, K99 and 987P) of enterotoxigenic Escherichia coli from neonatal and postweaning piglets with diarrhea in central China. Asian-Aust J Anim Sci. 2006, 19: 1342-1346. 10.5713/ajas.2006.1342.
Goetstouwers T, Van Poucke M, Coppieters W, Nguyen VU, Melkebeek V, Coddens A, Van Steendam K, Deforce D, Cox E, Peelman LJ: Refined Candidate Region for F4ab/ac enterotoxigenic Escherichia coli Susceptibility situated proximal to MUC13 in Pigs. PLoS One. 2014, 9 (8): e105013-10.1371/journal.pone.0105013.
Rampoldi A: Approach to the gene controlling the porcine receptor for Escherichia coli with fimbriae F4ab/F4ac and inheritance of the receptor for F4ad. ETH. PhD thesis. 2013, Institut für AgrarwissenschaftenInstitut für Agrarwissenschaften, ETH Zürich
Schroyen M, Stinckens A, Verhelst R, Niewold T, Buys N: The search for the gene mutations underlying enterotoxigenic Escherichia coli F4ab/ac susceptibility in pigs: a review. Vet Res. 2012, 43: 70-10.1186/1297-9716-43-70.
Koseki K, Wada T, Hosono M, Hata K, Yamaguchi K, Nitta K, Miyagi T: Human cytosolic sialidase NEU2-low general tissue expression but involvement in PC-3 prostate cancer cell survival. Biochem Biophys Res Commun. 2012, 428: 142-149. 10.1016/j.bbrc.2012.10.028.
Van den Broeck W, Cox E, Goddeeris BM: Receptor-specific binding of purified F4 isolated villi. Vet Microbiol. 1999, 68: 255-263. 10.1016/S0378-1135(99)00076-0.
Van den Broeck W, Cox E, Oudega B, Goddeeris BM: Receptor-dependent immune responses in pigs after oral immunization with F4 fimbriae. Infect Immunol. 1999, 67: 520-526.
Cox E, Houvenaghel A: Comparison of the in vitro adhesion of K88, K99, F41 and P987 positive Escherichia coli to intestinal villi of 4-week-old to 5-week-old pigs. Vet Microbiol. 1993, 34: 7-18. 10.1016/0378-1135(93)90003-P.
Jorgensen CB, Cirera S, Archibald AL, Anderson L, Fredholm M, Edfors-Lilja I: Porcine polymorphisms and methods for detecting them. International application published under the patent cooperation treaty (PCT). 2004, PCT/DK2003/000807 or WO2004/048606A2.
Li H, Li Y, Qiu X, Niu X, Liu Y, Zhang Q: Identification and screening of gene(s) related to susceptibility to enterotoxigenic Escherichia coli F4ab/ac in piglets. Asian-Austral J Anim. 2008, 21: 489-493. 10.5713/ajas.2008.70464.
Rasschaert R, Verdonck F, Goddeeris B, Duchateau L, Cox E: Screening of pigs resistant to F4 enterotoxigenic Escherichia coli (ETEC) infection. Vet Microbiol. 2007, 123: 249-253. 10.1016/j.vetmic.2007.02.017.
Gibbons RA, Sellwood R, Burrows M, Hunter PA: Inheritance of resistance to neonatal E. coli diarrhea in the pig: examination of the genetic system. Theor Appl Genet. 1977, 51: 65-70. 10.1007/BF00299479.
Hanada K: Sphingolipids in infectious diseases. Jpn J Infect Dis. 2005, 58: 131-148.
Taube S, Jiang M, Wobus CE: Glycosphingolipids as receptors for non-enveloped viruses. Viruses. 2010, 2: 1011-1049. 10.3390/v2041011.
Xu YH, Barnes S, Sun Y, Grabowski GA: Multi-system disorders of glycosphingolipid and ganglioside metabolism. J Lipid Res. 2010, 51: 1643-1675. 10.1194/jlr.R003996.
Bijlsma IGW, de Nijs A, van der Meer C, Frik JF: Different pig phenotypes affect adherence of Escherichia coli to jejunal brush borders by K88ab, K88ac, or K88ad antigen. Infect Immun. 1982, 37: 891-894.
Baker DR, Billey LO, Francis DH: Distribution of K88 Escherichia coli-adhesive and nonadhesive phenotypes among pigs of four breeds. Vet Microbiol. 1997, 54: 123-132. 10.1016/S0378-1135(96)01277-1.
Bonneau M, Duval-Iflah Y, Guerin G, Ollivier L, Renard C, Renjifo X: Aspects genetiques et microbiologiques de la colibacillose K88 chez le porc. Ann Rech Vet. 1990, 21: 302-303.
Li YH, Qiu XT, Li HJ, Zhang Q: Adhesive patterns of Escherichia coli F4 in piglets of three breeds. J Genet Genomics. 2007, 34: 591-599. 10.1016/S1673-8527(07)60067-8.
Yan X, Huang X, Ren J, Zou Z, Yang S, Ouyang J, Zeng W, Yang B, Xiao S, Huang L: Distribution of Escherichia coli F4 adhesion phenotypes in pigs of 15 Chinese and Western breeds and a White Duroc x Erhualian intercross. J Med Microbio. 2009, 58: 1112-1117. 10.1099/jmm.0.009803-0.
Van Poucke M, Vandesompele J, Mattheeuws M, Van Zeveren A, Peelman LJ: A dual fluorescent multiprobe assay for prion protein genotyping in sheep. BMC Infect Dis. 2005, 5: 13-10.1186/1471-2334-5-13.
Goetstouwers T, Van Poucke M, Nguyen VU, Melkebeek V, Coddens A, Dieter Deforce D, Cox E, Peelman LJ: F4ac-related expression analysis of the aminopeptidase N gene in pigs. J Anim Sci. 2014, 92: 1866-1873. 10.2527/jas.2013-7307.
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW: GenBank. Nucleic Acids Res. 2009, 37: D26-D31. 10.1093/nar/gkn723.
Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM: Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35: W71-W74. 10.1093/nar/gkm306.
Erkens T, Van Poucke M, Vandesompele J, Goossens K, Van Zeveren A, Peelman LJ: Development of a new set of reference genes for normalization of real-time RT-PCR data of porcine backfat and longissimus dorsi muscle, and evaluation with PPARGC1A. BMC Biotechnol. 2006, 6: 41-10.1186/1472-6750-6-41.
Acknowledgements
The authors wish to thank Linda Impe, Dominique Vander Donckt and Ruben Van Gansbeke for excellent technical assistance. This work was supported by an agricultural research grant of the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT Vlaanderen).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
TG performed the sample collection, carried out the genetic studies, performed data-analysis, and wrote the manuscript. MVP developed the protocols for the genetic studies, performed data-analysis, and participated in writing the manuscript. AC helped in designing the project and participated in reviewing the manuscript. VUN performed the sample collection and in vitro villous adhesion assay. LJP oversaw the project. All authors (TG, MVP, AC, VUN, VM DD, EC, and LJP) helped in designing the project, contributed in the data-analysis, reviewed and approved the manuscript.
Electronic supplementary material
12863_2014_103_MOESM1_ESM.docx
Additional file 1: Table S1.: Details of the investigated genes involved in the assembly of the F4 binding carbohydrate moiety of GSLs. (DOCX 17 KB)
12863_2014_103_MOESM2_ESM.docx
Additional file 2: Table S2.: Prevalence of the differential structural mutations in 11 investigated genes based on F4R type. (DOCX 57 KB)
12863_2014_103_MOESM3_ESM.docx
Additional file 3: Table S3.: Oligonucleotide sequences of primers with their specifications (ANPEP: NM_214277.1, ARSA: NM_213933.1, B4GALT6: XM_003127886.3, GAL3ST1: NM_001244429.1, GALC: NM_001243631.1, GBA: NM_001005730.1, GLA: NM_001177925.1, GLB1: AK230951.1, GLB1L: XM_001928375.3, NEU1: NM_001101822.1, NEU2: XM_003483766.2, UGCG: XM_001925267.5, UGT8: JQ650526). Table S4: PCR mix (10 μl) used in the semi-quantitative expression study via RT-PCR. Table S5: PCR program used in the semi-quantitative expression study via RT-PCR. (DOCX 25 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Goetstouwers, T., Van Poucke, M., Coddens, A. et al. Variation in 12 porcine genes involved in the carbohydrate moiety assembly of glycosphingolipids does not account for differential binding of F4 Escherichia coliand their fimbriae. BMC Genet 15, 103 (2014). https://doi.org/10.1186/s12863-014-0103-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12863-014-0103-x