Abstract
Background
In this work we have determined molecular signatures of oviduct epithelial and progenitor cells. We have proposed a panel of selected marker genes, which correspond with the phenotype of oviduct cells of a laying hen (Gallus gallus domesticus) and quail (Coturnix japonica). We demonstrated differences in characteristics of those cells, in tissue and in vitro, with respect to different anatomical and functional parts of the oviduct (infundibulum (INF), distal magnum (DM, and proximal magnum (PM)). The following gene expression signatures were studied: (1) oviduct markers (estrogen receptor 1, ovalbumin, and SPINK7 - ovomucoid), (2) epithelial markers (keratin 5, keratin 14, and occludin) and (3) stem-like/progenitor markers (CD44 glycoprotein, LGR5, Musashi-1, and sex determining region Y-box 9, Nanog homebox, OCT4/cPOUV gene encoding transcription factor POU5F3).
Results
In chicken, the expression of oviduct markers increased toward the proximal oviduct. Epithelial markers keratin14 and occludin were high in distal oviduct and decreased toward the proximal magnum. In quail oviduct tissue, the gene expression pattern of oviduct/epithelial markers was similar to chicken. The markers of progenitors/stemness in hen oviduct (Musashi-1 and CD44 glycoprotein) had the highest relative expression in the infundibulum and decreased toward the proximal magnum. In quail, we found significant expression of four progenitor markers (LGR5 gene, SRY sex determining region Y-box 9, OCT4/cPOUV gene, and CD44 glycoprotein) that were largely present in the distal oviduct. After in vitro culture of oviduct cells, the gene expression pattern has changed. High secretive potential of magnum-derived cells diminished by using decreased abundance of mRNA. On the other hand, chicken oviduct cells originating from the infundibulum gained ability to express OVM and OVAL. Epithelial character of the cells was maintained in vitro. Among progenitor markers, both hen and quail cells expressed high level of SOX9, LGR5 and Musashi-1.
Conclusion
Analysis of tissue material revealed gradual increase/decrease pattern in majority of the oviduct markers in both species. This pattern changed after the oviductal cells have been cultured in vitro. The results can provide molecular tools to validate the phenotype of in vitro biological models from reproductive tissue.
Similar content being viewed by others
Background
Avian oviduct in biomedical research
Avian species are excellent biological models in reproduction and tumorigenesis [1] as well as efficient source of secreting cells for use in bioreactors [2,3,4]. Both hen and quail oviduct cells secrete human therapeutic proteins after genetic modification [3]. Therefore the oviduct epithelium is a useful and fast in vitro model to test for the efficiency of viral [5] and nonviral genetic constructs [6] to study the modified secretome. Both quail and hen produce cellular substrates for the development of vaccines [7]. Genetic markers, including markers of stemness, are useful to identify mechanisms of malignant changes in a fallopian tube, because the somatic stem cells contribute to a population of tumor-initiating cells [8]. Recently, the knowledge about cell differentiation, physiology, and cancerogenic changes in avian oviduct has been extrapolated to women fallopian tube and uterine tract [9, 10]. However, in avian species, markers of stemness in oviduct cells have not been reported yet. There is a knowledge gap regarding distinctive features of the epithelial cells in in vitro conditions vs. their status in tissue, which limits full understanding and characterization of this cellular model. In this paper, we have made initial attempts to confirm progenitor molecular signatures in oviducts of laying hen (Gallus gallus domesticus) and quail (Coturnix japonica), both in tissue and in cultured oviduct epithelial cells (in vitro assay). We have addressed the following questions: What is the location of progenitor cells in avian oviduct tissue? What is an individual molecular characteristic of distal oviduct tissue compartments? Is this distinctive characteristic stable once the cells are plated in in vitro condition? Is the molecular pattern shared between these two model species (laying hen and quail) used for oviduct studies? Altogether, this study aims to provide a new understanding of molecular characteristic of oviduct epithelial cells in avian species.
Adult epithelial cells in the oviduct
In adult tissue, epithelial progenitor cells have limited potential to divide and they can develop only into few differentiated cell types. They express stem cell markers and can differentiate into epithelial cells with various phenotypes.
Mucous epithelium of an avian oviduct is composed of simple columnar cells equipped with cilia to move the ovum from distal to proximal oviduct and of nonciliated secreting cells. Both cell types require sustained renewal from the stem cell compartment and a high proliferation and maturation activity from the progenitor compartment. Those compartments are putatively based under the luminal epithelium as cellular niches [11]. In a mammalian fallopian tube, stem cells niches were tracked using antibodies and genetic markers and were found to be localized in the distal fallopian tube [8, 12]. In our earlier research, we determined faster proliferation of cultivated hen oviduct cells derived from infundibulum/distal magnum compared to the cells that were sourced from a proximal magnum. We have also determined that distal oviduct compartments were positively immunostained against CD44 and p63, which are known to be epithelial stem/progenitor markers [13]. Thereby, we have hypothesized that distal segments of avian oviduct contain progenitor gene expression signatures.
Genetic markers of distinctive signatures in avian oviduct epithelium
Characterization of oviduct cells using molecular markers for epithelial progenitors contributes to the understanding of differentiation and regeneration processes, which occur in the oviduct epithelium. As reported earlier, the self-renewal activity of cells in the fallopian tube occurs in its distal part [14, 15]. Thereby, in this paper, we have focused on distal parts of the oviduct (the closest to ovaries and abdomen) to follow the molecular characteristics of the cells in tissue and in vitro. We propose a panel of epithelial genetic markers to determine the progenitor/epithelial cell pattern in selected compartments of the oviduct (Fig. 1). In particular, we have aimed to reveal which of the avian oviduct compartments (infundibulum (INF), distal magnum (DM), or proximal magnum (PM)) carry known progenitor signatures.
A graphical representation of selected panel of epithelial genetic markers associated with oviduct cells. Three panels of epithelial genetic markers were proposed to provide a pattern of molecular signatures in the oviduct of hen and quail in 3 compartments: INF – infundibulum, DM – distal magnum, PM – proximal magnum. The first panel shown in the picture refers to stem-like markers: Nanog homebox (NANOG), octamer-binding protein 4 (OCT4/cPOUV) and sex determining region Y-box 9 (SOX9); and epithelial progenitor cells: cell surface glycoprotein CD44, leucine-rich repeat containing G protein-coupled receptor 5 (LGR5), and Musashi-1 (MSI-1). The second panel refers to epithelial cells: keratins KRT 5 and 14 and occludin (OCLN). The third panel refers to functional avian oviduct cells: estrogen receptor-1 (ESR1), ovalbumin (OVAL) and ovomucoid (OVM)
Methods
Isolation of the oviduct tissue
In this study, all the procedures involving experimental animals were approved by the Local Ethics Committee for Animal Research (http://lke.utp.edu.pl) located at the Faculty of Animal Breeding and Biology, UTP University of Science and Technology in Bydgoszcz (study approval reference number 35/2012, in accordance with the 2010_63_UE_PL Directive). The Hybrid Tetra SL laying hens (n = 6, 40 weeks old) were obtained from a commercial farm (Nowosc, Pradocin, Poland). Laying Japanese quails (n = 6, 10 weeks old) were obtained from a commercial producer (K. Drazek, Wyzne, Poland). All birds laid eggs at a daily rate and no hormonal stimulation was applied for this study. Immediately upon transportation, the animals were sacrificed by cervical dislocation. Only the oviducts after egg passage, with ovum position in a shell gland, were used for the experiments. Each oviduct was rinsed twice in tube filled out with 25 mL physiological buffered saline (PBS) w/o Mg, w/o Ca (Lonza Biosciences, Celllab, Warszawa, Poland), which was gently mixed with Penicillin-Streptomycin solution at 1:100 (v:v; Life Technologies, Warszawa, Poland).
In vitro culture of oviduct epithelial cells
The epithelial cells were isolated from the oviduct tissue using the methodology described earlier [16]. Immediately after tissue collection, three oviduct fragments were dissected: infundibulum, distal magnum, and proximal magnum, each 3 cm long. Each fragment was cleaned off the mesentery tissue and minced with a scalpel blade on a Petri dish. The minced fragments were digested in a solution of 1 mg/mL collagenase P (Sigma-Aldrich, Poznan, Poland) in Advanced Dulbecco’s Modified Eagles Medium-F12 (DMEM/F-12; Life Technologies, Warszawa, Poland) for 30 min at 37 °C, on a shaker. Due to the size of oviductal tubes, the amount of minced tissue was ~ 50% less in quail than in hen. Thus, adequately lower volumes of digestion solution were applied to process the quail oviduct tissue. The cells were counted manually using Neubauer hemocytometer and seeded at a density of 4 × 104 cells/cm2 into 25 cm2 vented BD type Primaria flasks (Becton Dickinson, Diag-med, Warszawa, Poland). The cells were incubated in 7% CO2 atmosphere at 37 °C. The oviduct cells were maintained in DMEM-F12 supplemented with 5% (v/v) fetal bovine serum (FBS; Life Technologies, cat. 16,140-063, batch No. 41G4541K, Warszawa, Poland), 1% (v/v) nonessential amino acids (Sigma-Aldrich, Poznan, Poland), 20 mM L-glutamine (Sigma-Aldrich, Poznan, Poland), 10 ng/mL human epidermal growth factor (human EGF; R&D Bioscience, Biokom, Janki, Poland), 1% (v/v) antibiotic – antimycotic solution (Life Technologies, Warszawa, Poland), 0.5 μg/mL hydrocortisone (Sigma Aldrich, Poznan, Poland) and 5 μg/mL insulin-transferrin-selenium (ITS; Sigma-Aldrich, Poznan, Poland). The viability and the proliferation of oviduct cells were measured by a real-time cell analyzer (RTCA) supplied by xCELLigence system (Roche Applied Science, Basel, Switzerland). The measurements of proliferating cells were conducted at 3.1 h intervals through 287 h post seeding, in accordance with the producer’s manual. The cells intended for sampling were cultivated for 5–7 days prior to harvesting and analysis. Every second day, the epithelial colonies were counted and photographed under an objective with phase contrast (Zeiss Axiovert 40) equipped with a digital camera (Canon EOS 600). The cells were harvested upon reaching 80% of growth confluence. Cultured oviduct epithelial cells were referred as chicken oviduct epithelial cells (COEC) or quail oviduct epithelial cells (QOEC) in further parts of this paper.
RNA isolation from oviduct tissue, COEC, and QOEC
RNA was isolated from three different sections of the oviduct tube (INF, DM, and PM) and cultivated oviduct cells, derived from the respective birds. For in vivo assay, INF, DM, and PM fragments, each 1 cm long, were cut off aseptically and put separately into Eppendorf tubes containing 3.0 mL RNAfix (EURx, Gdansk, Poland). Tissue samples were kept for 24 h at 4 °C and subsequently stored at − 20 °C until isolation of RNA. For RNA isolation from COEC, confluent cells were detached using Accutase® solution (A&E Life Sciences, Gentaur, Sopot, Poland) and centrifuged at 220×g for 5 min at room temperature (RT). Cell pellets were resuspended in 0.5 mL RNAfix (EURx, Gdansk, Poland) to preserve cells prior to RNA isolation. RNA was extracted using the universal RNA purification kit (EURx, Gdansk, Poland) according to manufacturer’s recommendation. RNA was quantified using spectrophotometry and RNA quality by gel electrophoresis.
RT-qPCR analysis
Reverse transcription was performed with Maxima First Strand cDNA synthesis kit for RT-qPCR (Thermo Scientific/Fermentas, Vilnius, Lithuania). cDNA was diluted to a final concentration of 70 ng/μL and stored at −20°C. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed in a total volume of 10 μL, which included Maxima SYBR Green qPCR Master Mix (Thermo Scientific/Fermentas, Vilnius, Lithuania), 1 μM of each primer (forward and reverse), and 2 μL of diluted cDNA (140 ng). Primer sequences (Table 1) were derived from the literature or designed with NCBI Primer Blast, based on cDNA reference sequences [17]. Thermal cycling was conducted in LightCycler II 480 (Roche Applied Science, Basel, Switzerland). qPCR thermal profile consisted of initial denaturation at 95 °C for 20 min, followed by 40 cycles of amplification including 15 s of denaturation at 95 °C, 20 s of annealing at 58 °C, and 20 s of elongation at 72 °C. After completion of the amplification reaction, a melting curve was generated to test for the specificity of RT-qPCR. For this purpose, the temperature was gradually increased to 98 °C with continuous fluorescence measurement.
Relative quantification of gene expression
Relative gene expression analysis was performed for each experimental group with ∆∆Ct method [18], using Ubiqutin C (UB) and β-actin (ACTB) as reference housekeeping genes. Geometric means of Ct value of both reference genes was used in calculations. For tested samples, ΔCt was calculated by subtracting mean Ct values of the reference genes from Ct values of the target gene. A base sample (calibrator) was defined by an origin different from the reproductive system. For in tissue study, muscle samples from the same birds were used. For in vitro study, the chicken macrophage-like cell line [19] was used as a calibrator. ΔΔCt was then calculated using the equation: ΔCt sample – ΔCt calibrator. Fold change of the gene expression was calculated as: R = 2–ΔΔCt.
Statistical analysis
RT-qPCR results were statistically analyzed using SAS Enterprise Guide 6.1 (SAS Institute, Cary, NC, USA). All tests were conducted on ΔCt values. First, Shapiro-Wilk test was used to assess the normality of data distribution. Then the significance of changes in the gene expression (in comparison to calibrator samples) was conducted by Student’s t-test (P < 0.05). Finally, multiple comparisons for all pairs (e.g., oviduct fragments or donor species) were performed with one-way ANOVA followed by Tukey’s HSD post hoc test. Standard error of the mean (SEM) was used as a parameter of variability within the group.
Results
Primary cultures of hen and quail oviduct epithelial cells
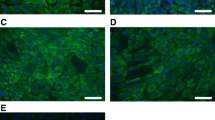
Cultivated oviduct cells of hen (COEC) and quail (QOEC) reached the confluence after 5–7 days after seeding. The COEC and QOEC isolated from the infundibulum region typically occurred as cellular spheres, which attached to the polystyrene culture vessel after 3 days post seeding and were consequently creating epithelial-like colonies, which spread on the surface of the culture vessel. Once the small epithelial colonies appeared beneath the spheres, they enter a high proliferation phase to rapidly form a confluent monolayer. Typical cultures from the infundibulum region were characterized by numerous compact epithelial islands, oval in shape, surrounded by elongated cells of mesenchymal or fibroblast-like phenotype (Fig. 2a, b). Epithelial cells isolated from the region of distal magnum (a transition region between infundibulum and proximal magnum) formed visible epithelial islands 3 days after seeding, which was sooner compared to the infundibulum region. In the case of distal magnum, spheres that formed epithelial-like colonies in vitro were half the size of those isolated from the infundibulum and about two times less colonies were initiated, compared to those from the infundibulum region on day 3 (Fig. 2c, d). In most cases, the epithelial colonies from the distal magnum proliferated fast and were ready for passage by 6–7 days after seeding. The cells from the region of proximal magnum usually did not form spheres in the beginning of the cultivation (Fig. 2e, f). Typically, in proximal magnum, the proliferating epithelial colonies were observed in 3–5 days post seeding. The microscopic observations of growing colonies were in line with the measurements acquired from the xCELLigence real-time cell monitoring system. The peak of the proliferation was determined for the cells from all oviduct compartments after 3 days post seeding: at 78.27 h for the INF part, at 79.05 h for the DM part, and at 79.05 h for the PM part. Then, the cell proliferation entered the plateau, which lasted 16.22 h for INF cells, 15.5 h for PM cells, and 10.85 h for DM cells. The cells from PM displayed larger morphology than cells from INF, and the shape of colonies was not compact, but oval and irregular. The confluent monolayer was heteromorphic, consisting of epithelial and fibroblast-like colonies (Fig. 2e, f). Motile cilia, which are characteristic for oviduct ciliated cells, were observed in the cultivated primary colonies, but only until the first passage. A movie file shows this in more detail (Additional file 1).
Phenotypes displayed by hen and quail oviduct cell colonies in vitro. a–b: confluent monolayers and visible spheres of colony-initiating cells isolated from the region of infundibulum neck (INF); magnification: × 100. c–d: confluent monolayers of epithelial cells isolated from distal magnum (DM), showing typical cobble-like morphology; magnification × 100. e–f: confluent epithelial monolayer, typically observed in cultivated cells that are originating from the oviduct magnum, showing mostly fibroblast-like morphology; magnification × 100. In each case, the cells were seeded at a density of 4 × 104 cells/cm2
Additional file 1: Visualization of a typical phenotype of cultivated oviductal epithelial cells. The recording of the cultivated oviduct epithelial cells allows one to follow the typical cobble-like structure of lining epithelial cells and the rotatory movement of cilia on the nonsecreting ciliated cells, which are coisolated with the secreting tubular gland cells. (MP4 12,229 kb)
Gene expression analysis
In this study, we have used three gene panels to characterize oviduct fragments of hen and quail, and the respective primary epithelial cell cultures that were derived from them. Those panels were comprised of oviduct (ESR1, OVAL, and OVM), epithelial (KRT5, KRT14, and OCLN), and stem-like/progenitor (LGR5, MSI1, SOX9, NANOG, and OCT4/cPOUV) gene expression signatures. Table 2 presents the overview of the gene function and the sequence similarity between a hen and a quail.
The overall gene expression of the markers analyzed in both species (hen and quail) and sample types (tissue and in vitro) is presented in Table 3. All twelve genes were expressed only in COEC. Ten out of twelve genes were expressed in oviduct tissues—sourced from both hen and quail. In the hen tissue, two progenitor markers (LGR5 and OCT4/cPOUV) were at a level too low to be detected. In the quail tissue, one epithelial marker (OCLN) and one progenitor marker (LGR5) were not detected. In QOEC, OVAL and OVM (oviduct markers) were not expressed as well as OCLN (epithelial marker). In both species, LGR5, a progenitor marker, was absent in the oviduct tissue, but then we detected it in the oviduct epithelial cell culture. OCLN was not expressed in quail oviduct—neither in the tissue, nor in the cell culture.
Characterization of gene expression signatures in hen and quail oviduct tissue
Hereby we have characterized gene expression profile in different parts (INF, DM, and PM) of hen and quail oviduct tissue (Fig. 3). In hen oviduct (Fig. 3a), the expression of oviduct markers (ESR1, OVAL and OVM) increased spatially, from distal to proximal part of the oviduct with a peak in PM (P < 0.05). Reversely, the expression of epithelial markers, KRT14 and OCLN, was high in INF and it decreased toward PM (P < 0.05). KRT5 was expressed at much lower level and only in INF (P < 0.05). As for progenitor markers, SOX9 was uniformly expressed at high level across all fragments of the oviduct in hen (P < 0.05). Expression of MSI1 and CD44 was the highest in INF and it gradually decreased toward PM (P < 0.05). Expression of NANOG was detected, but was not significant (P > 0.05).
Expression of oviduct, epithelial, and progenitor markers in different fragments of hen (a) and quail (b) oviduct tissue. Relative gene expression analysis was conducted with RT-qPCR method in three oviduct fragments: infundibulum (INF), distal magnum (DM) and proximal magnum (PM). Pairwise t-test was conducted to determine the significant modulation of the gene expression in the oviduct as compared to the external calibrator (breast muscle) (P < 0.05). An asterisk (*) indicates that the gene is differentially expressed, compared to the calibrator. Letters A, B, and C in brackets indicate results of one-way ANOVA multiple comparisons between different fragments of the oviduct (P < 0.05)
In quail oviduct (Fig. 3b), we had to use chicken primer sequences to show the expression pattern of OVAL, OVM, KRT5, KRT14, and OCT4/cPOUV. We have determined a similar pattern of the gene expression for two oviduct markers OVAL and OVM, which were expressed in all the studied oviduct compartments (INF, DM and PM). Whereas, oviduct marker for ESR1 was significantly expressed in INF and DM compartments of a quail oviduct (P < 0.05). Among epithelial markers, the expression of KRT14 and KRT5 was high and increased toward INF, but expression of OCLN did not reach the significance threshold. In quail, we found significant expression of as much as four progenitor markers (LGR5, OCT4/cPOUV, SOX9, and CD44) (P < 0.05). LGR5 and OCT4/cPOUV were most abundant in INF and DM compartments of the quail oviduct. Expression of NANOG was detected but it was not significant (P > 0.05).
Gene profiling of the gene expression signatures in COEC and QOEC
After having determined gene expression signatures in three specific fragments of chicken and quail oviducts, we have established the respective cell cultures, which were analyzed for the presence of the same markers. Results of the relative gene expression analysis in chicken and quail oviduct epithelial cells are presented in Fig. 4a. In COEC (Fig. 4a) only few markers were numerically and significantly upregulated, namely OVAL, OVM, KRT14, and SOX9 (P < 0.05). In the case of QOEC, we routinely found the abundance of ovalbumin in quail oviduct cell culture using antichicken OVA antibody and western blot detection. MSI1 was upregulated statistically (P < 0.05), though it did not have high numerical values of fold induction. In both, COEC and QOEC, OCT4/cPOUV was significantly downregulated (P < 0.05). We did not determine any significant differences between COEC derived from different fragments of the oviduct, apart from the expression of OCLN (epithelial marker) and LGR (progenitor marker) that was high in the INF compartment (P > 0.05).
Expression of oviduct, epithelial, and progenitor markers in chicken (a) and quail (b) oviduct epithelial cells. Relative gene expression analysis was performed with RT-qPCR method in three oviduct fragments: infundibulum (INF), distal magnum (DM) and proximal magnum (PM). Pairwise t-test was conducted to determine the significant modulation of the gene expression in the oviduct as compared to the external calibrator (breast muscle) (P < 0.05). An asterisk (*) indicates that the gene is differentially expressed, compared to the calibrator. Letters A, B, and C in brackets indicate results of one-way ANOVA multiple comparisons between different fragments of the oviduct (P < 0.05)
In QOEC, a similar significant expression, as in COEC, was found for progenitor markers SOX9 and MSI1 as well as epithelial marker KRT14. The remaining epithelial and stem-like/progenitor markers were significantly expressed in the cultivated quail cells derived from all studied compartments of oviduct (P > 0.05).
Discussion
Among avian species, laying hen (Gallus gallus domesticus) and Japanese quail (C. japonica) provide two excellent experimental oviduct models to study the immunology and reproductive biology [20]. Particular properties of oviductal cells include hormonal regulators as well as biosynthetic and secretive activity, which can be used for biomedical applications. Firstly, the secreting function of an oviduct epithelium makes it an ideal natural bioreactor to obtain human therapeutic proteins by using genetic manipulation of the oviduct secretome. The product is accumulated in the egg white and is easily harvested [3]. Secondly, both hen and quail are recognized to reflect the development and chemoprevention of spontaneous leiomyoma, also known as fibroids of the oviduct in relation to human cancer [21]. Thirdly, the development of new oviduct cell lines would allow selectively propagating and studying important pathogens including Campylobacter and Salmonella strains or influenza and Coronaviruses. Such cell lines offer new in vitro substrates for pathogens originating from a reproductive tract.
For this purpose, we have attempted to provide a utility set of molecular markers to characterize the avian oviduct tissue in hen and quail and in vitro-derived oviduct epithelial cell culture. For a quail, only 900 proteins are annotated in the existing UniProt databases. Thus, when a gap in quail database [22] limits the interpretation of a sequence, a relevant genomic alignment onto the chicken is performed [23]. Depending on the database used (ENSEMBL, NCBi, and/or UniProt), sequences of the genes selected for this study had 89%–100% similarity. Thereby, gene expression assays developed were comparable between both species.
In our study, all 12 analyzed genes were expressed in both hen and quail. In the first part, we have characterized gene expression signatures in three compartments of the oviduct tissue in hen and quail. The mRNA abundance of the oviduct markers (ESR1, OVAL, and OVM) increased toward proximal parts of the oviduct. Those differences between infundibulum and magnum compartments were significant only in hen earlier, but we have determined a clear numerical pattern also in quail. Such a pattern of the oviduct markers reflects physiological functions of distinct compartments, e.g., oocyte transport and sperm storage in the infundibulum vs. egg white protein production in the magnum. For this reason, ESR1, which encodes the estrogen receptor 1, whose major function is binding estradiol—a major sex hormone of laying birds, was expressed in all parts of the oviduct. On the other hand, OVAL and OVM, which encode major egg white proteins, were expressed only in the magnum. Such a pattern of the gene expression across the avian oviduct has been widely reported in the literature [24, 25] and it validates the functional setup of this experiment.
In a panel of epithelial markers characterized in tissue, we have determined a reverse pattern, i.e., decrease of mRNA abundance from distal toward proximal parts of oviduct, in particular of KRT14, which was strongly expressed in the infundibulum of both, hen and quail. KRT5 appeared to be more abundant in quail and OCLN was significantly expressed in chicken oviduct. Keratins encode for cytoskeletal proteins of highly proliferating basal epithelial cells [26]. Infundibulum is lined with ciliated epithelia, which are highly used by the frequent transportation of the oocyte and protein secretion. They require constant renewal from the basal epithelium, which is intensively proliferating. Strong induction of keratin genes is related with this function of the infundibulum. Previously, we have detected cytokeratins in chicken infundibulum by using immunohistochemistry technique, both in tissue and in vitro [6], which is in line with the results of the current study.
As for stem-like/progenitor markers analyzed in tissue, chicken expressed high mRNA abundance of CD44 and SOX9; moderate abundance of MSI1 and low of NANOG. OCT4/cPOUV and LGR5 were not expressed in the chicken oviduct tissue. In quail, we determined a high-fold induction of LGR5 and OCT4/cPOUV and a moderate abundance of CD44 and SOX9. CD44 is a cell surface glycoprotein and an established progenitor/stem-like cell marker in fallopian tube in mammals. CD44-positive cell population showed the capacity for clonal growth and differentiation into tubal epithelial cells, particularly in the distal region of the tube [15, 27]. We earlier showed a high immunochemical stain of CD44 in the distal oviduct of a hen [6]. SOX9 is a transcription factor in early epithelial lineage [28, 29]. It is involved in the organogenesis of different tissues and its main function is to maintain a population of undifferentiated somatic stem cells. SOX9 was recently announced as a novel cancer stem cell marker [30]. In our study, we consider this gene as a marker for precursor epithelial oviduct cells of avian species. Musashi-1 is expressed in intestinal crypts and human endometrium, where it maintains multipotent potential for epithelial cells emerging from Müllerian duct (precursor of oviduct in vertebrates) [31]. OCT4/cPOUV and NANOG are chicken stem cell markers [32, 33], while LGR5 is recognized as marker stem cells in tubal epithelia [14]. In our study, LGR5 and OCT4/cPOUV were detected at high level in quail oviduct. Overall, the pattern of expression of progenitor markers supports the designation of distal oviduct compartments as the source of progenitor epithelial cells.
After being transferred to in vitro conditions, phenotypes of COEC and QOEC have changed in some aspects. Secretive potential of magnum-derived cells was retained as reflected by the expression of OVM and OVAL in COEC and ESR1 in QOEC. Primary cultured cells, such as highly specialized oviduct epithelial cells, are prone to rapid differentiation in vitro. This way, they may easily lose their original phenotype, for example, the ability for protein secretion. On the other hand, INF-derived COEC gained secretive potential after being cultured in vitro, which was reflected by changing the downregulation of OVM and OVAL to upregulation of those genes as compared to donor tissues. Stimulation with estrogen was reported as necessary to maintain responsiveness of hen oviduct cells to this sex hormone [34]. However, in this experiment, neither the birds were stimulated with the estrogen prior to tissue harvesting, nor the cultivated cells were treated with estrogen, which might explain the lack of ESR-1 mRNA in the cultivated COEC. Epithelial character of both COEC and QOEC was maintained, especially in KRT14 (COEC) and other epithelial markers (QOEC) mRNA abundance.
Expression of progenitor markers of early epithelial lineage (SOX9, MSI1, and LGR5) in both oviduct epithelial cultures was determined. LGR5 was significantly upregulated in cultivated cells, and has been proven to mark the stem cells in murine oviduct/fimbria [14]. Precursor character of certain populations of cultured cells allowed for their proliferation and differentiation in vitro. INF-derived COEC gained gene expression signatures of oviduct secretive cells (OVM and OVAL). Population of progenitor cells is required for the establishment of a primary cell culture [35]. In our study, we have confirmed progenitor gene expression signatures in proliferating cultures. Based on the morphological assessment, a subpopulation of the cultured cells displayed epithelial character of ciliated and secreting cells. But there was also a large subpopulation of differentiated mesenchymal and fibroblast-like forms in both COEC and QOEC, after passaging. With these observations, a stable oviduct epithelial cell line could be probably established from both in vitro models, with the prior purification of progenitor cells from the heterogeneous starting cell populations.
Conclusion
In this study, we have characterized the expression of oviduct, epithelial, and stem/progenitor markers in the oviduct tissue and cell culture of two avian species, the hen and the quail. Analysis of the oviduct tissue and cultured cells allowed for characterizing the molecular makeup of those cells in tissue, in relation to the source of the oviduct compartment (infundibulum, distal magnum, and proximal magnum). Further analysis from in vitro-cultivated cells showed molecular pattern that was different from noncultivated oviduct cells. In conclusion, the analysis of tissue material revealed a gradual increase/decrease pattern in majority of the markers in both species. This pattern changed after those cells had been cultured in vitro. A progenitor marker, OCT4/cPOUV was strongly downregulated in both in vitro models, whereas the expression of SOX9 and the epithelial marker KRT14 were not changed compared to the calibrator (FC ~ 1). Cultivated hen cells (COEC) gained the expression of LGR5 progenitor marker, which could indicate a shift toward a more specific epithelial progenitor cell type. These results can contribute to further research on creating new biological models from reproductive tissue and the characterization required to develop new avian cell lines.
Abbreviations
- ACTB:
-
β-actin
- C. japonica :
-
Coturnix japonica
- CD44:
-
Cell surface glycoprotein
- COEC:
-
Chicken oviduct epithelial cells
- Ct:
-
Cycle threshold
- DM:
-
Distal magnum
- DMEM-F12:
-
Dulbecco’s Modified Eagles Medium-F12
- ESR1:
-
Estrogen receptor 1
- FBS:
-
Fetal bovine serum
- FC:
-
Fold Change
- G. gallus :
-
Gallus gallus
- GDP/GTP exchange factor VAV3:
-
Guanyl nucleotide exchange factor
- HMG:
-
High mobility group box transcription factor
- INF:
-
Infundibulum
- ITS:
-
Insulin-Transferrin-Selenium
- KRT14:
-
Keratin 14
- LGR5:
-
Leucine-rich repeat containing G protein-coupled receptor 5
- MSI-1:
-
Musashi-1
- NANOG:
-
Nanog homebox
- OCLN:
-
Occludin
- OCT4/cPOUV:
-
Octamer-binding protein 4
- OVAL:
-
Ovalbumin
- OVM:
-
Ovomucoid
- PM:
-
Proximal magnum
- QOEC:
-
Quail oviduct epithelial cells
- RT-qPCR:
-
Reverse transcription - quantitative polymerase chain reaction
- SEM:
-
Standard error of the mean
- SOX9:
-
Sex determining region Y-box 9
- UB:
-
Ubiquitin C
References
Trevino LS, Johnson PA. Estrogen receptor subtype expression is altered in the hen model of ovarian Cancer. J Mol Genet Med 2016;10. https://doi.org/10.4172/1747-0862.1000203.
Doran TJ, Cooper CA, Jenkins KA, Tizard MLV. Advances in genetic engineering of the avian genome: “Realising the promise.”. Transgenic Res. 2016;25:307–19.
Cao D, Wu H, Li Q, Sun Y, Liu T, Fei J, et al. Expression of recombinant human lysozyme in egg whites of transgenic hens. PLoS One 2015;10. https://doi.org/10.1371/journal.pone.0118626.
Zhang Z, Sun P, Yu F, Yan L, Yuan F, Zhang W, et al. Transgenic quail production by microinjection of lentiviral vector into the early embryo blood vessels. PLoS One. 2012;7:e50817.
Kwon SC, Choi JW, Jang H-J, Shin SS, Lee SK, Park TS, et al. Production of biofunctional recombinant human interleukin 1 receptor antagonist (rhIL1RN) from transgenic quail egg white. Biol Reprod. 2010;82:1057–64.
Stadnicka K, Bodnar M, Marszałek A, Bajek A, Drewa T, Płucienniczak G, et al. Efficient source of cells in proximal oviduct for testing non-viral expression constructs in the chicken bioreactor model and for other in vitro studies. Folia Biol (Krakow). 2016;64:37–46.
Genzel Y. Designing cell lines for viral vaccine production: where do we stand? Biotechnol J. 2015;10:728–40.
Garson K, Vanderhyden BC. Epithelial ovarian cancer stem cells: underlying complexity of a simple paradigm. Reproduction. 2015;149:R59–70.
Jo G, Lim W, Bae SM, Bazer FW, Song G. Avian SERPINB12 expression in the avian oviduct is regulated by estrogen and up-regulated in epithelial cell-derived ovarian carcinomas of laying hens. PLoS One. 2014;9:e99792.
Machado S, Bahr JM, Hales DB, Braundmeier AG, Quade BJ, Nowak R. Validation of the aging hen (Gallus gallus domesticus) as an animal model for uterine leiomyomas. Biol Reprod. 2012;87:86.
Kessler M, Zietlow R, Meyer TF. Adult Stem Cell Niches — Stem Cells in the Female Reproductive System. In: Wislet-Gendebien S, editor. Medicine- Stem Cell Research. InTech; 2014. https://doi.org/10.5772/58842. ISBN 978-953-51-1718-6.
Snegovskikh V, Mutlu L, Massasa E, Taylor HS. Identification of putative fallopian tube stem cells. Reprod Sci. 2014;21:1460–4.
Stadnicka K, Marszałek A, Kozłowska I, Walasik K, Bodnar M, Bajek A, et al. The expression of p63 and Ck HMW in magnum and infundibulum of Gallus domesticus oviduct. Folia Biol (Krakow). 2014;62:179–85.
Ng A, Tan S, Singh G, Rizk P, Swathi Y, Tan TZ, et al. Lgr5 marks stem/progenitor cells in ovary and tubal epithelia. Nat Cell Biol. 2014;16:745–57.
Paik DY, Janzen DM, Schafenacker AM, Velasco VS, Shung MS, Cheng D, et al. Stem-like epithelial cells are concentrated in the distal end of the fallopian tube: a site for injury and serous cancer initiation. Stem Cells. 2012;30:2487–97.
Kasperczyk K, Bajek A, Joachimiak R, Walasik K, Marszalek A, Drewa T, et al. In vitro optimization of the Gallus domesticus oviduct epithelial cells culture. Theriogenology. 2012;77:1834–45.
Ye J, Coulouris G, Zaretskaya I, Cutcutahe I, Rosen S, Madden TL. Primer- BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ÄÄCT method. Methods. 2001;25:402–8. https://doi.org/10.1006/meth.2001.1262.
Beug H, von Kirchbach A, Döderlein G, Conscience JF, Graf T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell. 1979;18:375–90.
Hosseini F, Hamsekolaei MHM, Moghaddam AKZ, Arabha H, Tohidifar SS. Normal ultrasonographic images of reproductive organs of female Japanese quails (Coturnix coturnix japonica): a laboratory animal model. Lab Anim 2016; https://doi.org/10.1177/0023677215625607.
Tuzcu M, Sahin N, Ozercan I, Seren S, Sahin K, Kucuk O. The effects of selenium supplementation on the spontaneously occurring fibroid tumors of oviduct, 8-hydroxy-2′-deoxyguanosine levels, and heat shock protein 70 response in Japanese quail. Nutr Cancer. 2010;62:495–500.
Kawahara-Miki R, Sano S, Nunome M, Shimmura T, Kuwayama T, Takahashi S, et al. Next-generation sequencing reveals genomic features in the Japanese quail. Genomics. 2013;101:345–53.
Kayang BB, Fillon V, Inoue-Murayama M, Miwa M, Leroux S, Fève K, et al. Integrated maps in quail (Coturnix japonica) confirm the high degree of synteny conservation with chicken (Gallus gallus) despite 35 million years of divergence. BMC Genomics. 2006;7:101.
Zhao J, Zhang Q, Jiao H, Wang X, Jiang M, Luo H, et al. Ovalbumin expression in the oviduct magnum of hens is related to the rate of egg laying and shows distinct stress-type-specific responses. J Anim Physiol Anim Nutr (Berl). 2016;100:876–83.
Nys Y, Gautron J, McKee MD, Garcia-Ruiz JM, Hincke MT. Biochemical and functional characterisation of eggshell matrix proteins in hens. Worlds Poult Sci J. 2001;57:401–13.
Couteaudier M, Trapp-Fragnet L, Auger N, Courvoisier K, Pain B, Denesvre C, et al. Derivation of keratinocytes from chicken embryonic stem cells: establishment and characterization of differentiated proliferative cell populations. Stem Cell Res. 2015;14:224–37.
Wang Y, Sacchetti A, van Dijk MR, van der Zee M, van der Horst PH, Joosten R, et al. Identification of quiescent, stem-like cells in the distal female reproductive tract. PLoS One. 2012;7:e40691.
Chue J, Smith CA. Sex determination and sexual differentiation in the avian model. FEBS J. 2011;278:1027–34.
Takada S, Ota J, Kansaku N, Yamashita H, Izumi T, Ishikawa M, et al. Nucleotide sequence and embryonic expression of quail and duck Sox9 genes. Gen Comp Endocrinol. 2006;145:208–13.
Kawai T, Yasuchika K, Ishii T, Miyauchi Y, Kojima H, Yamaoka R, et al. SOX9 is a novel cancer stem cell marker surrogated by osteopontin in human hepatocellular carcinoma. Sci Rep. 2016;6:e30489.
Lu X, Lin F, Fang H, Yang X, Qin L. Expression of a putative stem cell marker Musashi-1 in endometrium. Histol Histopathol. 2011;26:1127–33.
Lavial F, Acloque H, Bertocchini F, Macleod DJ, Boast S, Bachelard E, et al. The Oct4 homologue PouV and Nanog regulate pluripotency in chicken embryonic stem cells. Development. 2007;134:3549–63.
Lavial F, Pain B. Chicken embryonic stem cells as a non-mammalian embryonic stem cell model. Develop Growth Differ. 2010;52:101–14.
Jung JG, Park TS, Kim JN, Han BK, Lee SD, Song G, et al. Characterization and application of oviductal epithelial cells in vitro in Gallus domesticus. Biol Reprod. 2011;85:798–807.
Drewa T, Wolski Z, Pokrywka L, Debski R, Styczynski J. Progenitor cells are responsible for formation of human prostate epithelium primary cultures. Exp Oncol. 2008;30:148–52.
Caetano LC, Gennaro FGO, Coelho K, Araújo FM, Vila RA, Araújo A, et al. Differential expression of the MHM region and of sex-determining-related genes during gonadal development in chicken embryos. Genet Mol Res. 2014;13:838–49.
Sevane N, Bialade F, Velasco S, Rebolé A, Rodríguez ML, Ortiz LT, et al. Dietary inulin supplementation modifies significantly the liver transcriptomic profile of broiler chickens. PLoS One 2014;9. https://doi.org/10.1371/journal.pone.0098942.
De Boever S, Vangestel C, De Backer P, Croubels SSU. Identification and validation of housekeeping genes as internal control for gene expression in an intravenous LPS inflammation model in chickens. Vet Immunol Immunopathol. 2008;122:312–7.
Palmiter RD, Wrenn JT. Interaction of estrogen and progesterone in chick oviduct development. 3. Tubular gland cell cytodifferentiation. J Cell Biol. 1971;50:598–615.
Ichikawa K, Yamamoto I, Tsukada A, Saito N. cDNA cloning and mRNA expression of estrogen receptor alpha in Japanese quail. J Poult Sci. 2003;40:121–9.
Means AR, Comstock JP, O'Malley BW. Ovalbumin messenger RNA of chick oviduct: partial characterization, estrogen dependence, and translation in vitro. Proc Natl Acad Sci United States Am. 1972;69:1146–50.
Fertuck HC, Newstead JD. Fine structural observations on magnum mucosa in quail and hen oviducts. Z Zellforsch Mikrosk Anat. 1970;103:447–59.
Palmiter RD. Regulation of protein synthesis in Chick oviduct. I. Independent regulation of ovalbumin, conalbumin, ovomucoid, and lysozyme induction. J Biol Chem. 1972;247:6450–61.
Kato I, Schrode J, Kohr WJ, Laskowski M. Chicken ovomucoid: determination of its amino acid sequence, determination of the trypsin reactive site, and preparation of all three of its domains. Biochemistry. 1987;26:193–201.
Asai R, Okano H, Yasugi S. Correlation between Musashi-1 and c-hairy-1 expression and cell proliferation activity in the developing intestine and stomach of both chicken and mouse. Develop Growth Differ. 2005;47:501–10.
Auersperg N. The stem-cell profile of ovarian surface epithelium is reproduced in the oviductal fimbriae, with increased stem-cell marker density in distal parts of the fimbriae. Int J Gynecol Pathol. 2013;32:444–53.
Acknowledgements
Not applicable
Funding
This work was supported by the Polish National Science Center under grant agreement No. UMO-2011/03/N/NZ9/03814, the National Center for Research and Development under grant agreement No. PBS3/A8/30/2015 and the French-Polish project for collaborative research POLONIUM 2015–2016, granted to Bertrand Pain and Marek Bednarczyk. The research in this work was performed with equipment granted in the project “Implementation of the second phase of the Regional Center of Innovation” cofinanced by the European Regional Development Fund under the Regional Operational Program of Kujawsko-Pomorskie for the years 2007–2013.
Availability of data and materials
The data supporting the conclusions of this article are included within the article and its additional file. Any additional datasets which was used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
KS and AS made substantial contribution to conception and design of this manuscript. KS and AD acquired and interpreted the data reported. KS supervised the project and writing. KS, AS, and AD provided the original draft. MB BP and KS reviewed and edited the final manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Local Ethics Committee for Animal Research (http://lke.utp.edu.pl) located at the Faculty of Animal Breeding and Biology, UTP University of Science and Technology in Bydgoszcz (study approval reference number 35/2012).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Stadnicka, K., Sławińska, A., Dunisławska, A. et al. Molecular signatures of epithelial oviduct cells of a laying hen (Gallus gallus domesticus) and quail (Coturnix japonica). BMC Dev Biol 18, 9 (2018). https://doi.org/10.1186/s12861-018-0168-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12861-018-0168-2