Summary
The structural maintenance of chromosomes (SMC) proteins are essential for successful chromosome transmission during replication and segregation of the genome in all organisms. SMCs are generally present as single proteins in bacteria, and as at least six distinct proteins in eukaryotes. The proteins range in size from approximately 110 to 170 kDa, and each has five distinct domains: amino- and carboxy-terminal globular domains, which contain sequences characteristic of ATPases, two coiled-coil regions separating the terminal domains and a central flexible hinge. SMC proteins function together with other proteins in a range of chromosomal transactions, including chromosome condensation, sister-chromatid cohesion, recombination, DNA repair and epigenetic silencing of gene expression. Recent studies are beginning to decipher molecular details of how these processes are carried out.
Similar content being viewed by others
Gene organization and evolutionary history
Eukaryotes have at least six genes encoding structural maintenance of chromosomes (SMC) proteins, which are conserved from yeast to mammals; in several cases additional genes have been identified with either tissue-specific patterns of expression or additional functions [1,2]. The SMC genes were initially identified in genetic screens in the budding yeast Saccharomyces cerevisiae [3]. As their original name 'stability of minichromosomes' suggests, defects in SMC proteins destabilize chromosome segregation, and hence SMCs are in most cases essential for viability. SMC genes have been identified by both biochemical approaches in vertebrates and by further genetic screens in yeasts (S. cerevisiae and Schizosaccharomyces pombe), nematodes (Caenorhabditis elegans), insects (Drosophila melanogaster) and plants (Arabidopsis thaliana). The evolutionary conservation of SMC genes extends to prokaryotes; several prokaryotic species have a single gene, although there are two genes in Bacillus subtilis (encoding proteins that are 95% identical to each other) and a second potential SMC gene is found in Aquifex aeolicus (the two encoded SMC proteins are only 20% identical to each other). Interestingly, Escherichia coli does not have a gene encoding a bona fide SMC protein, but it does have a functionally conserved gene, mukB [4,5]. Details of gene names and map positions for the human SMC genes are shown in Table 1.
Phylogenetic analysis of SMC protein sequences places the six eukaryotic proteins in five families. The SMC5 and SMC6 proteins form a separate group less related to the eukaryotic families SMC1-SMC4 and have been grouped either together with the prokaryotic proteins in an 'ancestral' group [5], or as an independent group [1].
There is little direct information regarding the structure of SMC genes in higher eukaryote species, with the exception of murine SMC3, which comprises 31 exons spanning approximately 45 kb [6]. Draft human genomic sequences for human SMC3 (GenBank accession number NT_030081) and SMC4 (NT_005740) predict each gene to span approximately 35 kb, with 27 and 23 exons, respectively.
Characteristic structural features
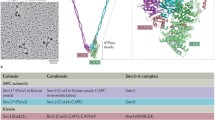
SMC proteins are large (approximately 110 to 170 kDa), and each is arranged into five recognizable domains (Figure 1). Rotary shadowing electron microscopy of several SMC dimers has shown that each has amino- and carboxy-terminal globular domains, separated by a rod with a central, flexible hinge [7]. Sequence analysis predicts the rod domain to be an extended coiled coil. The dimers are arranged in an antiparallel alignment. In eukaryotic cells, the proteins are found as heterodimers of SMC1 paired with SMC3, SMC2 with SMC4, and SMC5 with SMC6 (formerly known as Rad18) [1,8].
Structural features of the six human core SMC proteins. (a) Arrangement of domains in SMC proteins; see text for details. (b) Alignment of the residues in the amino-terminal domain of the six human SMC proteins, surrounding the 'Walker A' nucleotide-binding motif. Identical residues are shaded in yellow and conserved residues in green. Accession numbers of the sequences are listed in Table 1. (c) Alignment of the residues in the carboxy-terminal domain, surrounding the ABC signature and Walker B/DA-box motifs. Note the divergence in sequence in SMC5 and SMC6 compared to SMC1-SMC4.
Amino-acid sequence homology of SMC proteins between species is largely confined to the amino- and carboxy-terminal globular domains. The amino-terminal domain contains a 'Walker A' nucleotide-binding domain (GxxGxGKS/T, in the single-letter amino-acid code), which by mutational studies has been shown to be essential in several proteins. The carboxy-terminal domain contains a sequence (the DA-box) that resembles a 'Walker B' motif (ΦΦΦΦD, where Φ is any hydrophobic residue), and a motif with homology to the signature sequence of the ATP-binding cassette (ABC) family of ATPases. The sequence homology within the carboxy-terminal domain is relatively high within the SMC1-SMC4 group, whereas SMC5 and SMC6 show some divergence in both of these sequences (Figure 1). SMCs share not only sequence similarity but also structural similarity with ABC proteins, when the amino- and carboxy-terminal domains of two SMCs come together in a heterodimer. The crystal structure of a modified protein containing the amino and carboxyl termini of the Thermotoga maritima SMC shows structural homology to ABC proteins and to the Rad50 DNA-repair protein [9]. SMC proteins bind DNA, and for SMC1 the DNA-binding region has been mapped to the carboxy-terminal domain [10].
Localization and function
Consistent with their role in replication and chromosome and chromatid segregation, SMC proteins are found in all proliferating cells. B. subtilis SMC is localized to foci on the nucleoid and to the cell poles; it is not clear whether the SMC at the poles is present in functionally active complexes. The protein preferentially binds single-stranded DNA and aggregates DNA in the presence of ATP. The B. subtilis SMC gene is essential, and spores lacking SMC show defects in chromosome segregation during germination [4].
The six eukaryotic core SMCs (SMC1-SMC6) form functional complexes with other proteins. SMC1 and SMC3 are part of the cohesin complex, which contains two other proteins (sister-chromatid cohesion proteins Scc1 and Scc3) and is required for sister-chromatid cohesion during mitosis. The cohesin complex is loaded onto DNA during replication [2]. In yeast, the complexes remain bound to chromosomes until the metaphase-to-anaphase transition, at which stage proteolysis of the Scc1 subunit leads to the separation of paired sister chromatids. In Drosophila and human cells, the vast majority of cohesin disassociate from the chromosomes prior to mitosis, with the remaining cohesin being concentrated at the kinetochores. The kinetochore-bound fraction is also inactivated by proteolysis of the Scc1 subunit. The SMC1-SMC3 dimer also forms a recombination complex (RC-1) with DNA polymerase ε and ligase III [11]. Although SMC proteins are generally expressed in all proliferating cells, a meiotic SMC1 isoform was recently described in the mouse [12]. There are other components specific to meiotic cohesin, perhaps reflecting the large differences between chromosome segregation in meiosis and sister-chromatid segregation in mitosis.
SMC2 and SMC4 are part of the condensin complex, which contains three other proteins (CAP-D2/Cnd1, CAP-H/Cnd2 and CAP-G/Cnd3) and functions in the condensation of chromosomes during mitosis [1,2]. Condensin is localized along chromosomes during mitosis. Proteins related to SMC2 and SMC4 (MIX1 and DPY-27, respectively) in C. elegans have been shown to inactivate gene expression from the X chromosome in XX hermaphrodites to match that of XO males, a process known as dosage compensation [13,14]. A silencing function for the condensin subunit Barren (CAP-H/CND2) has also been described in Drosophila [15], which also uses topoisomerase II, another essential protein, for chromosome condensation.
Much less is known about the function of the SMC5 and SMC6 proteins. SMC6 corresponds to the Rad18 protein in S. pombe, with hypomorphic mutations resulting in defects in DNA repair and checkpoint signaling [8,16,17], and SMC6 also appears to be involved in recombination-based repair processes in yeast and Arabidopsis [18,19]. SMC5 and SMC6 genes are essential in yeast, although the essential function of the proteins is not known. SMC5 and SMC6 are also part of a multi-protein complex, but the identity of their binding partners is not known [20] - although rad18 mutants in S. pombe show genetic interactions with topoisomerase II and Brc1p, a BRCT-domain protein [16]. SMC6 is localized to chromatin in S. pombe and human cells, although in the latter it is not localized to mitotic chromosomes [8,16]. Murine SMC6 is extremely highly expressed in the testis and localizes to the sex chromosomes in late meiotic prophase [8].
Mechanism of action
The antiparallel arrangement of SMC dimers, with their flexible hinge, has led to the model that these molecules form V-shaped cross-linkages between different DNA molecules, in the case of cohesin, or within a single DNA molecule, in the case of condensin. Condensin complexes purified from mitotic Xenopus oocyte extracts induce DNA supercoiling in the presence of ATP, through the introduction of a global positive writhe [21]. Mitotic phosphorylation by the Cdc2 kinase of non-SMC subunits of the condensin complex is essential for these activities. Phosphorylation of histone H3 on serine 10, most likely by the Aurora kinases, seems be important for the recruitment of condensin onto mitotic chromosomes, given that condensin and phosphorylated histone H3 co-localize in immunostaining experiments. AKAP95, an anchoring protein that interacts with protein kinase A, has also been reported to recruit condensin [2].
Although the supercoiling of DNA by the condensin complex could account for a considerable degree of condensation, studies in Drosophila indicate that other activities are also required. Mutants with defects in gluon, the SMC4 homolog, have metaphase chromosomes that are the same length as wild-type controls, but these chromosomes do not have clearly resolved sister chromatids and are frequently broken during segregation [22]. So, although condensation and chromatid structure is disrupted, the shortening of the longitudinal axis is normal and is therefore presumably condensin-independent.
Recent studies have shown that the cohesin complex aggregates DNA. In contrast to the intramolecular knotting of DNA induced by the condensin complex, in the presence of topoisomerase II cohesin stimulates the formation of catenations in circular DNA [23]. Although the SMC dimers in the two complexes are very similar to each other, condensin and cohesin are differentially targeted to DNA to carry out distinct functions. Unlike bacterial SMCs, the eukaryotic proteins require interaction with the non-SMC components of the complexes for both localization and function.
Frontiers
To date, we understand little regarding the regulation of the SMC proteins by their non-SMC partners in multiprotein complexes. The SMC5-SMC6 complex is very poorly understood, both in terms of its components and its function, and this raises important questions. Furthermore, as topoisomerases are important players in chromosomal organization, the functional interactions between these proteins and the SMC-containing complexes will be important to decipher. Although details remain to be refined, it is already clear that the activity of these complexes must be coordinated for the ordered formation of mitotic chromosomes to occur. The study of the SMC protein family has considerably advanced our understanding of chromosome dynamics and will no doubt continue to do so in coming years.
References
Cobbe N, Heck MM: Review: SMCs in the world of chromosome biology - from prokaryotes to higher eukaryotes. J Struct Biol. 2000, 129: 123-143. 10.1006/jsbi.2000.4255. This review includes an extensive phylogenetic analysis of SMC family members, that places SMC5 and SMC6 (Rad18) in a group separate from prokaryotic SMCs.
Losada A, Hirano T: Shaping the metaphase chromosome: coordination of cohesion and condensation. Bioessays. 2001, 23: 924-935. 10.1002/bies.1133. The interplay between the functions of different SMC complexes in preparing chromosomes for mitosis is discussed in this review.
Strunnikov AV, Larionov VL, Koshland D: SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J Cell Biol. 1993, 123: 1635-1648. The identification of the first SMC protein.
Graumann PL: SMC proteins in bacteria: condensation motors for chromosome segregation?. Biochimie. 2001, 83: 53-59. 10.1016/S0300-9084(00)01218-9. This review presents a model for the function of prokaryotic SMC proteins throughout the cell cycle.
Jones S, Sgouros J: The cohesin complex: sequence homologies, interaction networks and shared motifs. Genome Biol. 2001, 2: research0009.1-0009.12. 10.1186/gb-2001-2-3-research0009. This paper presents a detailed bioinformatic analysis of the cohesin complex, and presents an evolutionary tree for the SMC proteins that groups SMC5 and SMC6 with the prokaryotic family members.
Ghiselli G, Siracusa LD, Iozzo RV: Complete cDNA cloning, genomic organization, chromosomal assignment, functional characterization of the promoter, and expression of the murine Bamacangene. J Biol Chem. 1999, 274: 17384-17393. 10.1074/jbc.274.24.17384. The only detailed information on genomic organization of a higher eukaryotic SMC gene is presented in this manuscript.
Melby TE, Ciampaglio CN, Briscoe G, Erickson HP: The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998, 142: 1595-1604. 10.1083/jcb.142.6.1595. Using electron microscopy, the first evidence for the structural arrangement of SMC and related polypeptides is presented.
Taylor EM, Moghraby JS, Lees JH, Smit B, Moens PB, Lehmann AR: Characterization of a novel human SMC heterodimer homologous to the Schizosaccharomyces pombeRad18/Spr18 complex. Mol Biol Cell. 2001, 12: 1583-1594. The first identification of human homologs of the yeast Rad18 and Spr18 proteins, together with the rationale to rename these SMC5 and SMC6.
Lowe J, Cordell SC, van den Ent F: Crystal structure of the SMC head domain: an ABC ATPase with 900 residues antiparallel coiled-coil inserted. J Mol Biol. 2001, 306: 25-35. 10.1006/jmbi.2000.4379. Structural data shows an ABC ATPase fold of the SMC heterodimer that is generated when the amino and carboxyl termini come together in a heterodimer.
Akhmedov AT, Frei C, Tsai-Pflugfelder M, Kemper B, Gasser SM, Jessberger R: Structural maintenance of chromosomes protein C-terminal domains bind preferentially to DNA with secondary structure. J Biol Chem. 1998, 273: 24088-24094. 10.1074/jbc.273.37.24088. Here, DNA-binding activity of SMC1 and SMC2 was first described, showing a preference for both AT-rich sequences and structures such as cruciform DNA.
Jessberger R, Riwar B, Baechtold H, Akhmedov AT: SMC proteins constitute two subunits of the mammalian recombination complex RC-1. EMBO J. 1996, 15: 4061-4068. The SMC1 and SMC3 proteins were identified as components of the RC-1 recombination complex, leading to the realization that SMC proteins have multiple different functions in addition to their function as part of the cohesin.
Revenkova E, Eijpe M, Heyting C, Gross B, Jessberger R: Novel meiosis-specific isoform of mammalian SMC1. Mol Cell Biol. 2001, 21: 6984-6998. 10.1128/MCB.21.20.6984-6998.2001. The existence of a meiotic SMC1 isoform may be important in the very different chromosome dynamics of meiosis I compared to mitosis.
Lieb JD, Albrecht MR, Chuang PT, Meyer BJ: MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell. 1998, 92: 265-277. See [15].
Chuang PT, Albertson DG, Meyer BJ: DPY-27: a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell. 1994, 79: 459-474. See [15].
Lupo R, Breiling A, Bianchi ME, Orlando V: Drosophila chromosome condensation proteins topoisomerase II and Barren colocalize with Polycomb and maintain Fab-7 PRE silencing. Mol Cell. 2001, 7: 127-136. This paper and [13,14] describe functions for SMC proteins in the epigenetic control of gene expression.
Verkade HM, Bugg SJ, Lindsay HD, Carr AM, O'Connell MJ: Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol Biol Cell. 1999, 10: 2905-2918. Defects in cell-cycle checkpoints are shown for the first time in an SMC mutant.
Lehmann AR, Walicka M, Grittiths DJF, Murray JM, Watts FZ, McCready S, Carr AM: The rad18 gene of Schizosaccharomyces pombedefines a new subgroup of the SMC superfamily involved in DNA repair. Mol Cell Biol. 1995, 15: 7067-7080. The initial identification of the SMC protein Rad18 (now renamed SMC6) in both fission and budding yeasts.
Hanin M, Mengiste T, Bogucki A, Paszkowski J: Elevated levels of intrachromosomal homologous recombination in Arabidopsis overexpressing the MIM gene. Plant J. 2000, 24: 183-189. 10.1046/j.1365-313X.2000.00867.x. See 19].
Mengiste T, Revenkova E, Bechtold N, Paszkowski J: An SMC-like protein is required for efficient homologous recombination in Arabidopsis. EMBO J. 1999, 18: 4505-4512. 10.1093/emboj/18.16.4505. This paper and [18] show a mutation in a gene closely related to SMC6 results in plants hypersensitive to DNA-damaging agents, most probably through a defect in recombination-based mechanisms of repair.
Fousteri MI, Lehmann AR: A novel SMC protein complex in Schizosaccharomyces pombecontains the Rad18 DNA repair protein. EMBO J. 2000, 19: 1691-1702. 10.1093/emboj/19.7.1691. Like the cohesin and condensin complexes, the Rad18 (SMC6) protein is shown to be part of a multi-protein complex.
Kimura K, Rybenkov VV, Crisona NJ, Hirano T, Cozzarelli NR: 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell. 1999, 98: 239-248. Biochemical evidence is presented to support a role for condensin in supercoiling and knotting of DNA strands.
Steffensen S, Coelho PA, Cobbe N, Vass S, Costa M, Hassan B, Prokopenko SN, Bellen H, Heck MM, Sunkel CE: A role for Drosophila SMC4 in the resolution of sister chromatids in mitosis. Curr Biol. 2001, 11: 295-307. 10.1016/S0960-9822(01)00096-3. The analysis of chromosomes in an SMC4 mutant (gluon) shows that the longitudinal axis of the chromosomes is shortened, as in wild-type cells, but the chromosomes are highly disorganized and unable to segregate.
Losada A, Hirano T: Intermolecular DNA interactions stimulated by the cohesin complex in vitro: implications for sister chromatid cohesion. Curr Biol. 2001, 11: 268-272. 10.1016/S0960-9822(01)00066-5. The intermolecular interactions between DNA strands mediated by the cohesin complex suggest a function of this complex in holding sister chromatids together. This is in contrast to condensin, which stimulates intramolecular knotting of DNA.
NCBI Map Viewer. The NCBI Map Viewer provides graphical displays of features on NCBI's assembly of human genomic sequence data. , [http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/map_search]
Acknowledgements
S.H.H. is a recipient of a Melbourne Research Scholarship. M.J.E.K. is a recipient of an Australian Postgraduate Award. M.J.O. is a Scholar of the Leukemia and Lymphoma Society.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Harvey, S.H., Krien, M.J. & O'Connell, M.J. Structural maintenance of chromosomes (SMC) proteins, a family of conserved ATPases. Genome Biol 3, reviews3003.1 (2002). https://doi.org/10.1186/gb-2002-3-2-reviews3003
Published:
DOI: https://doi.org/10.1186/gb-2002-3-2-reviews3003