Abstract
Background
Multidrug-resistant Plasmodium is of major concern today. Effective vaccines or successful applications of RNAi-based strategies for the treatment of malaria are currently unavailable. An unexplored area in the field of malaria research is the development of DNA-targeting drugs that can specifically interact with parasitic DNA and introduce deleterious changes, leading to loss of vital genome function and parasite death.
Presentation of the hypothesis
Advances in the development of zinc finger nuclease (ZFN) with engineered DNA recognition domains allow us to design and develop nuclease of high target sequence specificity with a mega recognition site that typically occurs only once in the genome. Moreover, cell-penetrating peptides (CPP) can cross the cell plasma membrane and deliver conjugated protein, nucleic acid, or any other cargo to the cytoplasm, nucleus, or mitochondria. This article proposes that a drug from the combination of the CPP and ZFN systems can effectively enter the intracellular parasite, introduce deleterious changes in its genome, and eliminate the parasite from the infected cells.
Testing the hypothesis
Availability of a DNA-binding motif for more than 45 triplets and its modular nature, with freedom to change number of fingers in a ZFN, makes development of customized ZFN against diverse target DNA sequence of any gene feasible. Since the Plasmodium genome is highly AT rich, there is considerable sequence site diversity even for the structurally and functionally conserved enzymes between Plasmodium and humans. CPP can be used to deliver ZFN to the intracellular nucleus of the parasite. Signal-peptide-based heterologous protein translocation to Plasmodium-infected RBCs (iRBCs) and different Plasmodium organelles have been achieved. With successful fusion of CPP with mitochondrial- and nuclear-targeting peptides, fusion of CPP with 1 more Plasmodium cell membrane translocation peptide seems achievable.
Implications of the hypothesis
Targeting of the Plasmodium genome using ZFN has great potential for the development of anti-malarial drugs. It allows the development of a single drug against all malarial infections, including multidrug-resistant strains. Availability of multiple ZFN target sites in a single gene will provide alternative drug target sites to combat the development of resistance in the future.
Similar content being viewed by others
Background
Malaria is the most devastating human parasitic infection. It threatens half of the world's population, killing more than 1 million people each year [1, 2]. Malaria species vary widely in epidemiology and clinical manifestation [3, 4]. No effective malaria vaccine is currently available, and drug resistance has been implicated in the spread and re-emergence of the disease [5–7]. Artemisinin drugs, an essential component of treatment for multidrug-resistant falciparum malaria, have recently shown signs of decreased efficacy in combination drug therapy [8–12].
With the availability of complete genome sequences of Plasmodium, humans and Anopheles-hosts of the parasite, development of novel effective anti malarial drugs was envisioned [13]. However, in spite of eclipse of more than one decade no significant advances have been made in the area of genomics based anti-malarial drug development. Plasmodium genome is highly AT rich as compared to human genome and this opens up opportunities for development of DNA-targeting drugs that can specifically interact with parasite DNA and introduce deleterious changes leading to loss of vital genome function and parasite death. Some DNA targeting drugs such as Aminoquinolines, azaterphenyl diamidines, adozelesin and bizelesin shows anti plasmodium activity, however the problem of cytotoxicity and potential threat of mutagenesis in human DNA is major limiting factor [14–16].
Presentation of the hypothesis
Advances in the development of zinc finger nuclease (ZFN) with engineered DNA recognition domains allow us to design and develop nuclease of high target sequence specificity [17–19]. In vivo application of ZFN with a single mega recognition sequence leads to target gene modification in Drosophila, zebra fish, mice, Arabidopsis, rice, tobacco, and other genomes [20–25], with a gene targeting frequency upto 80% of ZFN-transfected cells [26]. Cell-penetrating peptides (CPP) provide a novel mechanism for intracellular macromolecular delivery [27, 28], CPP are known to cross the cell plasma membrane in a nonspecific manner and deliver conjugated protein, nucleic acid, or any other cargo to the cytoplasm, nucleus, or mitochondria, depending on the additional signal sequences present [27, 29–33].
In this article it is believed that a drug from the combination of CPP and ZFN can effectively enter the intracellular parasite, introduce deleterious changes in its genome, and eliminate the parasite from infected cells. Preliminary studies of therapeutic application of ZFN-based drugs against HIV and hepatitis B virus further strengthen the prospects of ZFN-based anti-malarial drug development [26, 34]. ZFN mediated disruption of HIV co-receptor CCR5 in human CD4(+) T cells confers resistance to HIV-1 infection [22, 26, 35]. While ZFN therapeutic against hepatitis B virus intended to directly target and inactivate episomal DNA viral genomes [34]. In the present article, the use of ZFN to specifically target the Plasmodium genome with the aim to knock out vital parasite genes is proposed (Figure 1).
CPP-ZFN. A. Plasmodium-targeting peptide having signal peptides for infected RBC penetration, merozoite import, and organelle targeting, conjugated with Plasmodium specific zinc finger nuclease. B. It is hypothesized that the Plasmodium-targeting peptide will cross the membrane barriers of RBC and Plasmodium, and ZFN will introduce a double-strand break in the vital gene of the Plasmodium genome (a, b), leading to loss of gene function (c) and finally parasite death.
Key hypothesis/assumptions
-
1.
ZFN will specifically target the Plasmodium genome.
-
2.
ZFN can cross the plasma membrane barrier of the host and Plasmodium cell.
-
3.
DNA double-strand break will kill Plasmodium.
Testing of the hypothesis
Designing specific ZFN
ZFN can be designed specifically against any gene in any genome [21, 22, 36–38]. C2H2-type zinc finger proteins consist of an array of protein fingers stabilized by zinc ion, with each finger recognizing a specific triplet of DNA sequence [17]. Protein fingers specific for more than 45 triplets are known and have a modular nature, with freedom to change number of fingers, which makes the development of customized ZFP against diverse target DNA sequence feasible (Additional file 1). A combination of 6 fingers recognizes a DNA sequence of 18 nucleotides that have a probability of occurring after 418 nucleotides, much more than the human genome size (3000 MB). Hence, a ZFN designed against the Plasmodium genome have negligible probability of randomly occurring in the human genome. ZFN designing and validation tools, and CompoZr® custom ZFN designing, assembling, and validation service, reduce the time required for the development of ZFN against new targets [39–42].
Selection of ZFN target sites
Availability of complete genome sequences have helped identify different anti-malarial drug targets, such as histone deacetylase, aspartic proteases or plasmepsins, aminopeptidases, and the purine salvage enzyme hypoxanthine-xanthine-guanine phosphoribosyltransferase, within the parasite [43–46]. However, limited knowledge on the structure of most of these genes, and on the conservation of basic eukaryotic cellular machinery in Plasmodium and humans, hampers the conventional enzyme-targeting drug design procedures [47]. The Plasmodium genome is highly AT rich, in contrast to the human genome that is GC-rich; this provides considerable sequence (ZFN target) site diversity even for the structurally and functionally conserved enzymes between Plasmodium and humans. ZFN technology can utilize this sequence diversity and overcome the limitations of traditional drug-design approaches, which are confined to parasite-specific metabolic pathways. Moreover, ZFN can be designed against the target site conserved across different Plasmodium species (Additional file 2).
Delivery of ZFN to the target site
Discovery of cell-penetrating peptides provides a novel way of delivering therapeutic molecules to the target cell or organelles [27–29, 31, 48–51]. A major challenge in ZFN delivery is developing a peptide that can transfer conjugated ZFN through cell membranes of Plasmodium-infected RBC (iRBC), the parasite, parasitophorous vacuole, and its organelles (apicoplast, mitochondria, or nucleus).
Delivery to RBCs
CPP are generally considered to deliver cargo to cells in a nonspecific manner [51]. iRBCs have different surface proteins and charge that can be utilized for making strategies for specific ZFN delivery. As a matter of evidence, P1 peptides specifically target iRBCs [52], and DPT-sh1/sh2 CPP specifically recognize and deliver conjugated heterologous proteins to iRBCs [53].
Translocation from host cell cytoplasm to Plasmodium
Following CPP-mediated ZFN delivery to RBCs, the next challenge is to cross the parasite cell membrane and interact with its DNA. CPP-mediated protein delivery to different intracellular organelles such as the nucleus and mitochondria have been reported [54, 55]. Intracellular Plasmodium imports a number of proteins, such as heme, δ- aminolaevulinate (ALA), and peroxiredoxin 2, from the host cytoplasm [56–58], which could be a valuable source of signal sequences for translocation of ZFN from the host cytoplasm to the parasite.
Targeting to Plasmodium organelles
Once ZFN enters the Plasmodium, it has to be directed towards any of its organelles-apicoplast, mitochondria, or nucleus-which have their own genome. Use of the yeast GAL4 nuclear localization signal leads to translocation of green florescent protein (GFP) to Plasmodium nucleus and shows conservation of nuclear localization signal in Plasmodium and other eukaryotes [59]. GFP was fused with the N-terminal sequence of heat shock protein (PfHsp); PfHsp-GFP was targeted to Plasmodium mitochondria [60]; and apicoplast targeting was achieved by using the signal sequence of acyl carrier protein [61–63].
Availability of iRBCs penetrating peptide [53]; Plasmodium import proteins [56–58]; and Plasmodium apicoplast, mitochondria, and nucleus targeting signal sequences [59–63] provides building blocks for the development of a Plasmodium-targeting, multiple-membrane-traversing peptide. Following the successful fusion of CPP with mitochondrial- and nuclear-targeting peptides [54, 64, 65], fusion of CPP with 1 more Plasmodium cell membrane translocation peptide appears to be achievable.
Hydrolysis of double-stranded DNA
A ZFN-induced double-strand break (DSB) triggers either of the 2 DNA repair processes: error-prone non-homologous end joining (NHEJ) and error-proof homology-dependent repair (HDR). Genetic and biochemical evidence support the hypothesis that NHEJ and HR are 2 independent and competing mechanisms for DSB repair in diploid organisms[66, 67]. Being in the haploid stage in humans, HDR must be absent in Plasmodium and the error-prone repair mechanism of NHEJ often results in localized mutations due to deletion and/or insertion of short sequences at the DSB site, thereby resulting in disruption of functional gene expression in Plasmodium. Moreover, AT-specific DNA alkylating drugs have previously shown anti P. falciparum activity and recent advances in ZFN directed multiple DSB resulting in chromosomal deletions further strengthens the prospects of DNA targeting anti malaria drug [68–70].
Experimental validation of ZFN therapeutic efficacy
An in silico designed and in vitro validated ZFN is to be evaluated for bioavailability, potency, in vivo localization and their specificity for parasite over human DNA. A microfluorimetric method using PicoGreen® can be used for assessing susceptibility of parasites to CPP-ZFN compounds [71, 72]. Since all ZFN consist of nuclease domain of FokI restriction enzyme so FokI antisera can be used for immuno-localization and quantification of ZFN [73]. Alternatively ZFN are designed to have an additional small protein tag, such as Flag or His tag and subsequently detected by fusion tag specific antibodies [74]. The quantification of DSB repair loci induced by ZFN provide genotoxicity assay, to test any off target effect of parasite specific ZFN over human genome [75, 76].
Implications of the hypothesis
The major advantage of ZFN technology is that it enormously increases the number of drug targets because it can utilize the vast sequence diversity among structurally and functionally conserved enzymes of human and Plasmodium proteins. Prolonged use of any drug forces the evolution of drug-resistant parasite strains, and ZFN would not be an exception. Another advantage of ZFN technology is that it allows convenient development of new ZFN against other target sites (18 bp) in the same gene. The resulting new drug will be effective even against resistant strains, providing ample alternatives for drug resistance management.
Unlike common drugs that directly inhibit the target protein, ZFN technology is focused on inhibiting the synthesis of a functional target protein; thus, its effect will be slower than inhibitor drugs. This constraint may not be a concern as ZFN utilizes the difference between human and Plasmodium gene sequences, so every vital gene becomes a potential drug target. Selection of genomic locus coding for short half-life will greatly enhance the response time of ZFN-based drugs.
Availability of newer drug targets to virtually all Plasmodium genes provides the opportunity to find conserved sites in vital genes and develop a single drug against all malarial infections, including multidrug-resistant strains, for worldwide use. CPP-mediated protein delivery is an established method that can be used for delivery of the newly designed ZFN. CPP-ZFN promises to be a safe and sustainable drug for malaria intervention.
Conflict of interests
The authors declare that they have no competing interests.
References
Phillips RS: Current status of malaria and potential for control. Clin Microbiol Rev. 2001, 14: 208-226. 10.1128/CMR.14.1.208-226.2001.
Vitoria M, Granich R, Gilks CF, Gunneberg C, Hosseini M, Were W, Raviglione M, De Cock KM: The global fight against HIV/AIDS, tuberculosis, and malaria: current status and future perspectives. Am J Clin Pathol. 2009, 131: 844-848. 10.1309/AJCP5XHDB1PNAEYT.
Prugnolle F, Durand P, Neel C, Ollomo B, Ayala FJ, Arnathau C, Etienne L, Mpoudi-Ngole E, Nkoghe D, Leroy E, Delaporte E, Peeters M, Renaud F: African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc Natl Acad Sci USA. 2010, 107: 1458-1463. 10.1073/pnas.0914440107.
Jeffares DC, Pain A, Berry A, Cox AV, Stalker J, Ingle CE, Thomas A, Quail MA, Siebenthall K, Uhlemann AC, Kyes S, Krishna S, Newbold C, Dermitzakis ET, Berriman M: Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet. 2007, 39: 120-125. 10.1038/ng1931.
Escalante AA, Smith DL, Kim Y: The dynamics of mutations associated with anti-malarial drug resistance in Plasmodium falciparum. Trends Parasitol. 2009, 25: 557-563. 10.1016/j.pt.2009.09.008.
Guerin PJ, Bates SJ, Sibley CH: Global resistance surveillance: ensuring antimalarial efficacy in the future. Curr Opin Infect Dis. 2009, 22: 593-600. 10.1097/QCO.0b013e328332c4a7.
Witkowski B, Berry A, Benoit-Vical F: Resistance to antimalarial compounds: methods and applications. Drug Resist Updat. 2009, 12: 42-50. 10.1016/j.drup.2009.01.001.
Denis MB, Tsuyuoka R, Lim P, Lindegardh N, Yi P, Top SN, Socheat D, Fandeur T, Annerberg A, Christophel EM, Ringwald P: Efficacy of artemether-lumefantrine for the treatment of uncomplicated falciparum malaria in northwest Cambodia. Trop Med Int Health. 2006, 11: 1800-1807. 10.1111/j.1365-3156.2006.01739.x.
Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM: Evidence of artemisinin-resistant malaria in western Cambodia. NEJM. 2008, 359: 2619-2620. 10.1056/NEJMc0805011.
Denis MB, Tsuyuoka R, Poravuth Y, Narann TS, Seila S, Lim C, Incardona S, Lim P, Sem R, Socheat D, Christophel EM, Ringwald P: Surveillance of the efficacy of artesunate and mefloquine combination for the treatment of uncomplicated falciparum malaria in Cambodia. Trop Med Int Health. 2006, 11: 1360-1366. 10.1111/j.1365-3156.2006.01690.x.
Alker AP, Lim P, Sem R, Shah NK, Yi P, Bouth DM, Tsuyuoka R, Maguire JD, Fandeur T, Ariey F, Wongsrichanalai C, Meshnick SR: Pfmdr1 and in vivo resistance to artesunate-mefloquine in falciparum malaria on the Cambodian-Thai border. Am J Trop Med Hyg. 2007, 76: 641-647.
Dondorp AM, Yeung S, White L, Nguon C, Day NP, Socheat D, von Seidlein L: Artemisinin resistance: current status and scenarios for containment. Nat Rev Microbiol. 2010, 8: 272-280. 10.1038/nrmicro2385.
Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B: Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002, 419: 498-511. 10.1038/nature01097.
Wenzel NI, Chavain N, Wang Y, Friebolin W, Maes L, Pradines B, Lanzer M, Yardley V, Brun R, Herold-Mende C, Biot C, ToÌth K, Davioud-Charvet E: Antimalarial versus Cytotoxic properties of dual drugs derived from 4-aminoquinolines and mannich bases: interaction with DNA. J Med Chem. 2010, 53: 3214-3226. 10.1021/jm9018383.
Hu L, Arafa RK, Ismail MA, Patel A, Munde M, Wilson WD, Wenzler T, Brun R, Boykin DW: Synthesis and activity of azaterphenyl diamidines against Trypanosoma brucei rhodesiense and Plasmodium falciparum. Bioorg Med Chem. 2009, 17: 6651-6658. 10.1016/j.bmc.2009.07.080.
Woynarowski JM, Krugliak M, Ginsburg H: Pharmacogenomic analyses of targeting the AT-rich malaria parasite genome with AT-specific alkylating drugs. Mol Biochem Parasitol. 2007, 154: 70-81. 10.1016/j.molbiopara.2007.04.009.
Carroll D: Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 2008, 15: 1463-1468. 10.1038/gt.2008.145.
Kandavelou K, Mani M, Durai S, Chandrasegaran S: "Magic" scissors for genome surgery. Nat Biotechnol. 2005, 23: 686-687. 10.1038/nbt0605-686.
Cathomen T, Joung JK: Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008, 16: 1200-1207. 10.1038/mt.2008.114.
Foley JE, Maeder ML, Pearlberg J, Joung JK, Peterson RT, Yeh JR: Targeted mutagenesis in zebrafish using customized zinc-finger nucleases. Nat Protoc. 2009, 4: 1855-1867. 10.1038/nprot.2009.209.
Kandavelou K, Chandrasegaran S: Custom-designed molecular scissors for site-specific manipulation of the plant and mammalian genomes. Methods Mol Biol. 2009, 544: 617-636. full_text.
Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS: Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009, 19: 1279-1288. 10.1101/gr.089417.108.
Lee HJ, Kim E, Kim JS, Radecke S, Radecke F, Cathomen T, Schwarz K: Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2009, 20: 81-89. 10.1101/gr.099747.109.
Remy S, Tesson L, Menoret S, Usal C, Scharenberg AM, Anegon I, Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, Rebar EJ, Gregory PD, Klug A, Collingwood TN, Wu J, Kandavelou K, Chandrasegaran S, Mani M, Kandavelou K, Dy FJ, Durai S, Chandrasegaran S: Zinc-finger nucleases: a powerful tool for genetic engineering of animals. Transgenic Res. 2009, 26: 26-
Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, Choi VM, Rock JM, Wu YY, Katibah GE, Zhifang G, McCaskill D, Simpson MA, Blakeslee B, Greenwalt SA, Butler HJ, Hinkley SJ, Zhang L, Rebar EJ, Gregory PD, Urnov FD: Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009, 459: 437-441. 10.1038/nature07992.
Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, Guschin DY, Rupniewski I, Waite AJ, Carpenito C, Carroll RG, Orange JS, Urnov FD, Rebar EJ, Ando D, Gregory PD, Riley JL, Holmes MC, June CH: Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008, 26: 808-816. 10.1038/nbt1410.
Sawant R, Torchilin V, Raagel H, Saalik P, Pooga M, Munst B, Patsch C, Edenhofer F, Gitton Y, Tibaldi L, Dupont E, Levi G, Joliot A, Edenhofer F, Meade BR, Dowdy SF, Gump JM, Dowdy SF, Meade BR, Dowdy SF, Dong X, Wang JN, Huang YZ, Guo LY, Kong X, Murriel CL, Dowdy SF: Intracellular transduction using cell-penetrating peptides. Mol Biosyst. 2009, 6: 628-640. 10.1039/b916297f.
Gump JM, Dowdy SF, Meade BR, Dowdy SF, Dong X, Wang JN, Huang YZ, Guo LY, Kong X, Murriel CL, Dowdy SF: TAT transduction: the molecular mechanism and therapeutic prospects. Trends Mol Med. 2007, 13: 443-448. 10.1016/j.molmed.2007.08.002.
Ward B, Seal BL, Brophy CM, Panitch A, Sawant R, Torchilin V, Raagel H, Saalik P, Pooga M, Munst B, Patsch C, Edenhofer F, Gitton Y, Tibaldi L, Dupont E, Levi G, Joliot A, Edenhofer F, Meade BR, Dowdy SF, Gump JM, Dowdy SF, Meade BR, Dowdy SF, Dong X, Wang JN, Huang YZ, Guo LY, Kong X, Murriel CL, Dowdy SF: Design of a bioactive cell-penetrating peptide: when a transduction domain does more than transduce. J Pept Sci. 2009, 15: 668-674. 10.1002/psc.1168.
Gitton Y, Tibaldi L, Dupont E, Levi G, Joliot A, Edenhofer F, Meade BR, Dowdy SF, Gump JM, Dowdy SF, Meade BR, Dowdy SF, Dong X, Wang JN, Huang YZ, Guo LY, Kong X, Murriel CL, Dowdy SF: Efficient CPP-mediated Cre protein delivery to developing and adult CNS tissues. BMC Biotechnol. 2009, 9: 40-10.1186/1472-6750-9-40.
Fonseca SB, Pereira MP, Kelley SO: Recent advances in the use of cell-penetrating peptides for medical and biological applications. Adv Drug Deliv Rev. 2009, 61: 953-964. 10.1016/j.addr.2009.06.001.
Meade BR, Dowdy SF, Gump JM, Dowdy SF, Meade BR, Dowdy SF, Dong X, Wang JN, Huang YZ, Guo LY, Kong X, Murriel CL, Dowdy SF: Enhancing the cellular uptake of siRNA duplexes following noncovalent packaging with protein transduction domain peptides. Adv Drug Deliv Rev. 2008, 60: 530-536. 10.1016/j.addr.2007.10.004.
Meade BR, Dowdy SF, Dong X, Wang JN, Huang YZ, Guo LY, Kong X, Murriel CL, Dowdy SF: Exogenous siRNA delivery using peptide transduction domains/cell penetrating peptides. Adv Drug Deliv Rev. 2007, 59: 134-140. 10.1016/j.addr.2007.03.004.
Cradick TJ, Keck K, Bradshaw S, Jamieson AC, McCaffrey AP: Zinc-finger nucleases as a novel therapeutic strategy for targeting hepatitis b virus DNAs. Mol Ther. 2010, 2010: 16-
Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, Cannon PM: Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotech. 2010, 28: 839-847. 10.1038/nbt.1663.
Cai CQ, Doyon Y, Ainley WM, Miller JC, Dekelver RC, Moehle EA, Rock JM, Lee YL, Garrison R, Schulenberg L, Blue R, Worden A, Baker L, Faraji F, Zhang L, Holmes MC, Rebar EJ, Collingwood TN, Rubin-Wilson B, Gregory PD, Urnov FD, Petolino JF: Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol Biol. 2009, 69: 699-709. 10.1007/s11103-008-9449-7.
Pruett-Miller SM, Connelly JP, Maeder ML, Joung JK, Porteus MH: Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol Ther. 2008, 16: 707-717. 10.1038/mt.2008.20.
Wu J, Kandavelou K, Chandrasegaran S, Mashimo T, Takizawa A, Voigt B, Yoshimi K, Hiai H, Kuramoto T, Serikawa T: Custom-designed zinc finger nucleases: what is next?. Cell Mol Life Sci. 2007, 64: 2933-2944. 10.1007/s00018-007-7206-8.
Jayakanthan M, Muthukumaran J, Chandrasekar S, Chawla K, Punetha A, Sundar D: ZifBASE: a database of zinc finger proteins and associated resources. BMC Genomics. 2009, 10: 421-10.1186/1471-2164-10-421.
Fu F, Sander JD, Maeder M, Thibodeau-Beganny S, Joung JK, Dobbs D, Miller L, Voytas DF: Zinc Finger Database (ZiFDB): a repository for information on C2H2 zinc fingers and engineered zinc-finger arrays. Nucleic Acids Res. 2009, D279-283. 10.1093/nar/gkn606. 37 Database
Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D: Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 2007, W599-605. 10.1093/nar/gkm349. 35 Web Server
Mandell JG, Barbas CF: Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006, W516-523. 10.1093/nar/gkl209. 34 Web Server
Na-Bangchang K, Karbwang J: Current status of malaria chemotherapy and the role of pharmacology in antimalarial drug research and development. Fundam Clin Pharmacol. 2009, 23: 387-409. 10.1111/j.1472-8206.2009.00709.x.
Choi SR, Mukherjee P, Avery MA: The fight against drug-resistant malaria: novel plasmodial targets and antimalarial drugs. Curr Med Chem. 2008, 15: 161-171. 10.2174/092986708783330575.
Jana S, Paliwal J: Novel molecular targets for antimalarial chemotherapy. Int J Antimicrob Agents. 2007, 30: 4-10. 10.1016/j.ijantimicag.2007.01.002.
Gardiner DL, Skinner-Adams TS, Brown CL, Andrews KT, Stack CM, McCarthy JS, Dalton JP, Trenholme KR: Plasmodium falciparum: new molecular targets with potential for antimalarial drug development. Expert Rev Anti Infect Ther. 2009, 7: 1087-1098. 10.1586/eri.09.93.
Dharia NV, Chatterjee A, Winzeler EA: Genomics and systems biology in malaria drug discovery. Curr Opin Investig Drugs. 11: 131-138.
Juliano RL, Alam R, Dixit V, Kang HM: Cell-targeting and cell-penetrating peptides for delivery of therapeutic and imaging agents. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2009, 1: 324-335. 10.1002/wnan.4.
Vives E, Schmidt J, Pelegrin A: Cell-penetrating and cell-targeting peptides in drug delivery. Biochim Biophys Acta. 2008, 1786: 126-138.
Morris MC, Deshayes S, Heitz F, Divita G: Cell-penetrating peptides: from molecular mechanisms to therapeutics. Biol Cell. 2008, 100: 201-217. 10.1042/BC20070116.
Torchilin VP: Tat peptide-mediated intracellular delivery of pharmaceutical nanocarriers. Adv Drug Deliv Rev. 2008, 60: 548-558. 10.1016/j.addr.2007.10.008.
Eda K, Eda S, Sherman IW: Identification of peptides targeting the surface of plasmodium falciparum-infected erythrocytes using a phage display peptide library. Am J Trop Med Hyg. 2004, 71: 190-195.
Guergnon J, Dessauge F, Dominguez V, Viallet J, Bonnefoy S, Yuste VJ, Mercereau-Puijalon O, Cayla X, Rebollo A, Susin SA, Bost PE, Garcia A: Use of penetrating peptides interacting with PP1/PP2A proteins as a general approach for a drug phosphatase technology. Mol Pharmacol. 2006, 69: 1115-1124. 10.1124/mol.105.019364.
Vyas PM, Payne RM: TAT opens the door. Mol Ther. 2008, 16: 647-648. 10.1038/mt.2008.24.
Flierl A, Jackson C, Cottrell B, Murdock D, Seibel P, Wallace DC: Targeted delivery of DNA to the mitochondrial compartment via import sequence-conjugated peptide nucleic acid. Mol Ther. 2003, 7: 550-557. 10.1016/S1525-0016(03)00037-6.
Bonday ZQ, Taketani S, Gupta PD, Padmanaban G, Bonday ZQ, Dhanasekaran S, Rangarajan PN, Padmanaban G: Heme biosynthesis by the malarial parasite. Import of delta-aminolevulinate dehydrase from the host red cell. J Biol Chem. 1997, 272: 21839-21846. 10.1074/jbc.272.35.21839.
Bonday ZQ, Dhanasekaran S, Rangarajan PN, Padmanaban G: Import of host delta-aminolevulinate dehydratase into the malarial parasite: identification of a new drug target. Nat Med. 2000, 6: 898-903. 10.1038/78659.
Koncarevic S, Rohrbach P, Deponte M, Krohne G, Prieto JH, Yates J, Rahlfs S, Becker K: The malarial parasite Plasmodium falciparum imports the human protein peroxiredoxin 2 for peroxide detoxification. Proc Natl Acad Sci USA. 2009, 106: 13323-13328. 10.1073/pnas.0905387106.
Wittayacom K, Uthaipibull C, Kumpornsin K, Tinikul R, Kochakarn T, Songprakhon P, Chookajorn T: A nuclear targeting system in Plasmodium falciparum. Malar J. 9: 126-10.1186/1475-2875-9-126.
Sato S, Rangachari K, Wilson RJ: Targeting GFP to the malarial mitochondrion. Mol Biochem Parasitol. 2003, 130 (2): 155-158. 10.1016/S0166-6851(03)00166-X.
Stanway RR, Witt T, Zobiak B, Aepfelbacher M, Heussler VT: GFP-targeting allows visualization of the apicoplast throughout the life cycle of live malaria parasites. Biol Cell. 2009, 101: 415-430. 10.1042/BC20080202. 415 p following 430.
Tonkin CJ, Kalanon M, McFadden GI: Protein targeting to the malaria parasite plastid. Traffic. 2008, 9: 166-175.
Tonkin CJ, Foth BJ, Ralph SA, Struck N, Cowman AF, McFadden GI: Evolution of malaria parasite plastid targeting sequences. Proc Natl Acad Sci USA. 2008, 105: 4781-4785. 10.1073/pnas.0707827105.
Horton KL, Stewart KM, Fonseca SB, Guo Q, Kelley SO: Mitochondria-penetrating peptides. Chem Biol. 2008, 15: 375-382. 10.1016/j.chembiol.2008.03.015.
Yoshikawa T, Sugita T, Mukai Y, Yamanada N, Nagano K, Nabeshi H, Yoshioka Y, Nakagawa S, Abe Y, Kamada H, Tsunoda S, Tsutsumi Y: Organelle-targeted delivery of biological macromolecules using the protein transduction domain: potential applications for peptide aptamer delivery into the nucleus. J Mol Biol. 2008, 380: 777-782. 10.1016/j.jmb.2008.05.047.
Essers J, van Steeg H, de Wit J, Swagemakers SM, Vermeij M, Hoeijmakers JH, Kanaar R: Homologous and non-homologous recombination differentially affect DNA damage repair in mice. Embo J. 2000, 19: 1703-1710. 10.1093/emboj/19.7.1703.
Liang F, Han M, Romanienko PJ, Jasin M: Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci USA. 1998, 95: 5172-5177. 10.1073/pnas.95.9.5172.
Woynarowski JM, Krugliak M, Ginsburg H: Pharmacogenomic analyses of targeting the AT-rich malaria parasite genome with AT-specific alkylating drugs. Molecular and Biochemical Parasitology. 2007, 154: 70-81. 10.1016/j.molbiopara.2007.04.009.
Lee HJ, Kim E, Kim J-S: Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Research. 2010, 20: 81-89. 10.1101/gr.099747.109.
Sollu C, Pars K, Cornu TI, Thibodeau-Beganny S, Maeder ML, Joung JK, Heilbronn R, Cathomen T: Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion. Nucl Acids Res. 2010, gkq720.
Quashie NB, de Koning HP, Ranford-Cartwright LC: An improved and highly sensitive microfluorimetric method for assessing susceptibility of Plasmodium falciparum to antimalarial drugs in vitro. Malar J. 2006, 5: 95-10.1186/1475-2875-5-95.
Corbett Y, Herrera L, Gonzalez J, Cubilla L, Capson TL, Coley PD, Kursar TA, Romero LI, Ortega-Barria E: A novel DNA-based microfluorimetric method to evaluate antimalarial drug activity. Am J Trop Med Hyg. 2004, 70: 119-124.
Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D: Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics. 2006, 172: 2391-2403. 10.1534/genetics.105.052829.
Pruett-Miller SM, Reading DW, Porter SN, Porteus MH: Attenuation of zinc finger nuclease toxicity by small-molecule regulation of protein levels. PLoS Genet. 2009, 5: e1000376-10.1371/journal.pgen.1000376.
Cornu TI, Cathomen T: Quantification of zinc finger nuclease-associated toxicity. Methods Mol Biol. 2010, 649: 237-245. full_text.
Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ: A rapid and general assay for monitoring endogenous gene modification. Meth Mol Biol. 2010, 649: 247-256. full_text.
Acknowledgements
The authors are thankful to Drs. Jane Carlton and Steven Sullivan, New York University Langone Medical Center, New York, for their critical reading of the manuscript and for providing valuable suggestions that increased scientific content and presentation of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Authors' contributions
VN: conceived the idea. AV, SS, and VN: involved in intellectual discussions, formulated the hypothesis, and wrote the manuscript. SS and VN: created graphical presentations and designed ZFN against the target site. All authors read and approved the final manuscript.
Electronic supplementary material
12936_2010_1339_MOESM1_ESM.TIFF
Additional file 1: Designing of target-site-specific zinc finger nuclease (ZFN) through modular assembly. With the target site specific ZFP designing by incorporating zinc finger helices, linkers, N and C-terminal fixed sequences, ZFP binding to target site can be validated by gel shift assay. As ZFP do not have nuclease activity it does not cut the target sequence. Incorporation of nuclease domain of FokI constitutes the functional ZFN that can be validated by in vitro digestion of DNA with target sequence. (TIFF 3 MB)
12936_2010_1339_MOESM2_ESM.TIFF
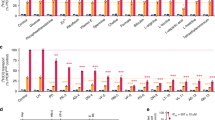
Additional file 2: Designing of zinc finger nuclease (ZFN) against a conserved site in lactate dehydrogenase genes of P. falciparum and P. vivax. Two ZFN designed on opposite strands will introduce a nick in the spacer region, leading to a double-strand break. ELISA data of multi-target specificity assay for all triplets, black bars represent target oligonucleotides, while white bars represent oligonucleotide pools with a particular 5' nucleotide. The height of each bar represents the relative specificity of the protein for each target [42]. (TIFF 2 MB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Nain, V., Sahi, S. & Verma, A. CPP-ZFN: A potential DNA-targeting anti-malarial drug. Malar J 9, 258 (2010). https://doi.org/10.1186/1475-2875-9-258
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-9-258