Abstract
Background
The sporulation of aerial hyphae of Streptomyces coelicolor is a complex developmental process. Only a limited number of the genes involved in this intriguing morphological differentiation programme are known, including some key regulatory genes. The aim of this study was to expand our knowledge of the gene repertoire involved in S. coelicolor sporulation.
Results
We report a DNA microarray-based investigation of developmentally controlled gene expression in S. coelicolor. By comparing global transcription patterns of the wild-type parent and two mutants lacking key regulators of aerial hyphal sporulation, we found a total of 114 genes that had significantly different expression in at least one of the two mutants compared to the wild-type during sporulation. A whiA mutant showed the largest effects on gene expression, while only a few genes were specifically affected by whiH mutation. Seven new sporulation loci were investigated in more detail with respect to expression patterns and mutant phenotypes. These included SCO7449-7451 that affect spore pigment biogenesis; SCO1773-1774 that encode an L-alanine dehydrogenase and a regulator-like protein and are required for maturation of spores; SCO3857 that encodes a protein highly similar to a nosiheptide resistance regulator and affects spore maturation; and four additional loci (SCO4421, SCO4157, SCO0934, SCO1195) that show developmental regulation but no overt mutant phenotype. Furthermore, we describe a new promoter-probe vector that takes advantage of the red fluorescent protein mCherry as a reporter of cell type-specific promoter activity.
Conclusion
Aerial hyphal sporulation in S. coelicolor is a technically challenging process for global transcriptomic investigations since it occurs only as a small fraction of the colony biomass and is not highly synchronized. Here we show that by comparing a wild-type to mutants lacking regulators that are specifically affecting processes in aerial hypha, it is possible to identify previously unknown genes with important roles in sporulation. The transcriptomic data reported here should also serve as a basis for identification of further developmentally important genes in future functional studies.
Similar content being viewed by others
Background
The developmental life cycle of Streptomyces coelicolor belongs to the most complex among prokaryotes. After a spore has germinated and grown out into a vegetative mycelial network, multicellular developmental processes lead to both the onset of secondary metabolism and the emergence of specialised reproductive hyphae that form an aerial mycelium on the surface of colonies (reviewed in [1, 2]). The initiation of development involves both sensing of nutritional stimuli and complex extracellular signalling, including quorum sensing, extracellular proteases, and other putative signals (see e.g. [3–5]). The formation of aerial hyphae depends on a series of mostly regulatory genes that have been designated bld since they are required for the emergence of the hairy aerial mycelium on the colony surface. The regulatory networks governed by these genes are only partially understood, but are gradually being revealed [4, 6, 7].
The subsequent development of the aerial hyphae into spores can be blocked at different stages by mutating critical genes. Many mutations of this type give rise to a white aerial mycelium due to a failure to produce the grey spore pigment. Isolation of such whi mutants was the basis for identifying central regulatory genes that direct sporulation in aerial hyphae (for recent reviews, see [1, 4]). A major challenge in Streptomyces developmental biology is now to decipher how these regulators are acting to control the physiological and cell cycle-related processes involved in producing the mature spores, including modulation of cell division, cell wall assembly, chromosome replication, and nucleoid partitioning and condensation. The accompanying physiological responses include for example the cell type-specific accumulation and utilisation of glycogen and trehalose, and the synthesis of a polyketide spore pigment. The biosynthetic genes for the pigment are found in the whiE gene cluster, and the expression of this cluster depends on the regulatory whi genes, although the direct regulator is still unknown [8, 9].
The identified regulatory whi genes that are required for the early stages of sporulation in aerial hyphae appear to fall into two major and converging pathways [1]. The RNA polymerase sigma factor σWhiG is required for the initiation of spore formation in S. coelicolor and controls two other regulatory genes, whiI encoding a response regulator and whiH encoding a GntR-family protein [10–13]. Genetic analyses show that whiG mutations block progression of differentiation at an early stage of apparently undifferentiated aerial hyphae in S. coelicolor, and whiG mutations are epistatic on both whiI and whiH[14, 15]. The phenotypes of whiI and whiH mutants differ in that whiI mutants do not form sporulation septa and do not show pronounced nucleoid condensation, while whiH mutants are able to convert the apical cells of some aerial hyphae into spore-like fragments with condensed nucleoids and occasional sporulation septa [12, 13, 15]. WhiH is autoregulatory and binds to its own promoter region [16], while WhiI (C-terminal fragment) binds to one independent target promoter (for inoRA) [17, 18]. However, no other direct targets for WhiH or WhiI have been reported. A parallel pathway seems to be controlled by whiA and whiB. Orthologues of whiA are found in most Gram-positive bacteria and their gene products have a bipartite structure consisting of a domain similar to a class of homing endonucleases combined with a DNA-binding domain in the shape of a helix-turn-helix motif [19–21]. S. coelicolor WhiA is so far reported to bind directly to its own promoter and to a sporulation-induced promoter controlling the parAB genes [22]. WhiB is the founding member of the actinomycete-specific Wbl (WhiB-like) family of FeS-cluster proteins that appear to act in transcription control, although functions ascribed to Wbl proteins have been controversial [4, 23–26]. Disruption of whiA or whiB arrests sporulation at a very early stage, and mutant phenotypes of the two are indistinguishable [15, 19, 23].
The two converging pathways that depend on whiG-whiI/whiH and whiA/whiB, respectively, are required for controlling most aspects of the conversion of aerial hyphae into spores. However, very few direct targets are known for these central regulatory whi genes, and overall it seems like only a small subset of genes involved in aerial hyphal sporulation have been identified. In order to find further genes that are developmentally regulated in S. coelicolor and involved in the differentiation of aerial hyphae to spores, we have carried out a DNA microarray-based transcriptome analysis. The experiment was designed to identify genes that are up-regulated during development of the wild-type parent but are not up-regulated in derivative strains bearing mutations in either whiA or whiH, representing the two abovementioned sporulation-specific pathways. For a subset of the genes that were identified as developmentally regulated and specifically affected by whiA and/or whiH, we have confirmed expression patterns using real-time qRT-PCR, S1 nuclease mapping, and reporter gene fusions, and constructed and analysed deletion mutants. This has identified a set of previously unknown developmentally regulated promoters and sporulation genes that encode different types of regulators, a protease, an L-alanine dehydrogenase, and proteins related to spore pigment biogenesis.
Results and discussion
Transcriptional analysis of whiA- and whiH-dependent gene expression during development of S. coelicolor
A developing S. coelicolor colony is a complex mixture of cells at different developmental stages, and the sporulating aerial mycelium constitutes only a fraction of the total colony biomass. In order to identify genes that are specifically changed in sporulating aerial hyphae, we have therefore compared the pattern of gene expression in the wild-type strain M145 to those in two developmental mutants lacking the regulatory genes whiA or whiH (strains J2401 and J2408, respectively). Disruption of these genes imposes specific blocks or defects at an early stage of aerial hyphal sporulation without overtly affecting any other cell type. Mycelium was harvested after 18, 36 and 48 h of growth, in the case of the wild-type strain representing colonies consisting of vegetative mycelium only, colonies covered by a developing aerial mycelium, and colonies turning grey due to abundant production of spores, respectively. RNA was isolated from four independent cultures of each strain and used to generate Cy3- and Cy5-labelled cDNA. For each time point, pairs of Cy3- and Cy5-labelled cDNA of wild-type and one of the two mutants were co-hybridized on DNA microarrays according to a balanced block design [27], with a total of four array hybridizations for each comparison (Figure 1). In addition to the comparisons of wild-type vs whi mutant samples, cDNA of wild-type samples from 36 and 48 h were hybridized to the 18 h sample to reveal genes changing during development of the wild-type strain (Figure 1). In total, eight different class comparisons were conducted.
Schematic view of the experimental design used to compare the transcriptomes of whiA and whiH mutants to that of the wild type M145 strain. A18 refers to whiA mutant cDNA from 18 h growth, A36 is whiA cDNA from 36 h, A48 from 48 h. W refers to wild type strain M145 and H to the whiH mutant. At 18 h, samples consisted mainly of vegetative mycelium (Veg), while aerial hyphae formation (AHF) was seen at 36 h, and abundant spores (Sp) were produced at 48 h in the wild-type cultures.
Only considering differences in expression with a Benjamini-Hochberg corrected p-value < 0.05 as significant [28], we found a total of 285 genes differentially expressed in at least one of the 8 class comparisons analyzed (Additional file 1: Table S1). 114 of them (Figure 2) had significantly different levels of transcription in at least one time point of the whiA or whiH mutant compared to the wild-type, and the following discussion concerns these 114 genes only. Most of the significant effects of the whiA and whiH mutations could be seen at the latest time point, and no gene with significant change of expression between mutant and the parent was detected at 18 h. This is consistent with our initial assumption that whiA and whiH specifically affect gene expression in sporulating aerial mycelium. Only a few genes were significantly affected by whiA or whiH disruption at 36 h, including seven in the whiA and six in the whiH strain. At 48 h, 103 genes were changed significantly in the whiA strain compared to the parent (29 with higher expression and 74 with lower expression than in the wild-type), while only 25 where changed in the whiH mutant (7 with higher expression and 18 with lower expression than in the wild-type). The change in expression level among the 114 differentially expressed genes ranged from +1.5 to +6.7 fold for the genes overexpressed in the mutants as compared to the wild type, and -1.5 to -24.7 fold for the under-expressed ones. 44 out of the 114 genes showed more than 2 fold change of the expression level. Of the 114 genes that were affected by whi mutations, 13 were previously known to be involved in the differentiation processes or to be closely related to such genes (Additional file 2: Figure S1).
Hierarchical clustering of the 114 genes that were found to be significantly differentially expressed in at least one comparison between a mutant and the wild-type parent strain. A18, A36, and A48 refer to comparison of whiA mutant cDNA to wild-type cDNA prepared from developmental time points 18 h, 36 h, and 48 h, respectively. H refers to similar comparisons of whiH to wild-type at the given time points, and wt36 and wt48 refer to comparison of cDNA from wild-type strain at 36 h and 48 h, respectively, compared to the 18 h sample (as illustrated in Figure 1). Colour-coded expression values (log2) are shown, where blue indicates lower expression and yellow indicates higher expression in mutant compared to wild-type (or in wild-type 36 h or 48 h sample compared to 18 h sample). Grey boxes indicate comparisons for which there is no expression value since not all four arrays showed at least one good spot.
Both hierarchical clustering of the 114 differentially expressed genes according to their expression profiles (Figure 2) and grouping in a Venn diagram (Figure 3) indicated four dominant patterns. Genes with increased expression in a mutant compared to wild-type parent fell into two distinct subgroups at 48 h, showing overexpression only in the whiA or the whiH mutant, respectively. Only one gene was significantly overexpressed in both mutants (SCO3113). Among the genes with down-regulated expression in at least one mutant, the majority showed increased expression during development of the wild-type strain, further supporting the notion that these genes are related to the sporulation process. Two main subgroups were recognised, with one being affected by both whiA and whiH, and the other only affected by whiA (Figures 2 and 3). Figure 3 indicates three genes that may specifically depend on whiH for developmental up-regulation, but closer examination of the data showed that all three (SCO0654, SCO6240, SCO7588) have decreased expression in the whiA mutant also, albeit with a Benjamini-Hochberg corrected p-value >0.05 (Additional file 1: Table S1). Thus, all of the genes that were down-regulated in the whiH strain appeared to be also down-regulated in the whiA mutant, while another group only depended on whiA and not whiH. This is consistent with whiA mutations giving a more complete block of sporulation than whiH mutations [15], and it suggests that there may be very few genes that specifically depend on whiH for expression.
Venn diagrams showing the distributions of differentially expressed genes (with a Benjamini-Hochberg corrected p-value <0.05) among samples from the whiA (A) and whiH (H) mutants and different time points (36 h and 48 h). “Down-regulated” refers to genes with expression value significantly lower in the mutant sample compared to the respective wild-type sample, and “up-regulated” refers to genes with significantly higher expression in mutant compared to wild-type.
To further verify the microarray data, we have used qRT-PCR to test expression of 17 genes with decreased expression in one or both mutants (putative sporulation-induced genes). This overall expression pattern was confirmed for several genes, with eleven out of the 17 tested genes showing a significantly lower expression in the whiA mutant compared to the wildtype at at least one of the two sporulation time points 36 h and 48 h (Additional file 2: Figure S2). Thus, a large fraction of this group are developmentally regulated genes correctly identified by the array analysis. Further investigations of several of these genes are described in the following sections.
For the genes that appeared overexpressed in the whiH mutant, i.e. that were putative candidates for being repressed by WhiH, six genes were tested by qRT-PCR. Five appeared to be false positives and only one had its microarray expression profile confirmed by qRT-PCR experiments (Additional file 2: Figure S3). This is the previously described gene eshB (SCO5249) encoding a putative cyclic nucleotide-binding protein [29]. The qRT-PCR indicated higher eshB expression during development of the whiH mutant compared to the parent strain. In an S1 nuclease protection assay (Additional file 2: Figure S4), the eshB promoter was found to be similarly up-regulated during development in both the parent and the whiH mutant, and the level of transcript was only 1.4-fold higher in the mutant at the 36 h time point and not different from wildtype at 48 h (after normalisation to the hrdB promoter as internal control). Also the eshB paralogoue eshA (SCO7699) [29] was significantly up-regulated in the whiH mutant according to the arrays (Additional file 2: Figure S3), but S1 nuclease protection assays showed that eshA is strongly up-regulated during developmental in both strains, with only subtle difference in mRNA level between the whiH mutant and the wild-type (Additional file 2: Figure S4). Overall, our analyses did not reveal any clear candidates for repression by the WhiH transcription factor.
Analysis of expression and mutant phenotypes of new sporulation genes
We have specifically investigated seven potential sporulation loci emerging from the microarray analysis (Figure 4). Expression of these loci has been monitored using qRT-PCR (Figure 5), S1 nuclease mapping (Figure 6), and promoter fusions to a reporter gene encoding the fluorescent protein mCherry (Figure 7 and Table 1). For the latter experiments, we constructed a new vector, pKF210, used this to construct “promoter probe” fusions, and introduced them into Streptomyces strains (described in Materials and Methods). Furthermore, deletion mutants have been constructed for these seven loci and examined to detect phenotypes associated with sporulation and maturation of spores. The tested features were colony appearance and pigmentation on MS agar; appearance of aerial hyphae and spores in phase-contrast microscopy; and heat resistance of spores. One additional sporulation-induced locus that was discovered through this study has already been reported, namely hupS (SCO5556) encoding a nucleoid-associated HU-like protein that influences nucleoid structure and spore maturation [30].
Gene organization along the chromosome of S. coelicolor for the seven new sporulation loci that are described in this paper. (A-G) Genes for which deletion mutants have been constructed are drawn in black. The immediately surrounding genes are shown in grey. DNA fragments used for complementation of deletion mutants are indicated by a line for loci SCO7449-7451 (F) and SCO1774-1773 (G). For the SCO1774-1773 locus, the results of a semi-quantitative RT-PCR assay are summarized (H). The data are shown in Additional file 2: Figure S5. The presence of different kinds of transcripts in strain M145 is indicated for RNA prepared from vegetative and sporulating mycelium (H). The primer pairs used for RT-PCR (specified in Additional file 1: Table S1) are designated 1, 2, 3, and drawn as arrows. Detection of a transcript is indicated with a plus (+) and the absence with a minus (-). The relative amount of the PCR product is indicated by one or two plus signs. The indicated sporulation induced P1774 promoter (G) was identified by S1 nuclease mapping (see Figure 6A).
Quantitative real-time RT-PCR assays of selected genes. Specific primer pairs were used to amplify SCO0934, SCO1195, SCO1773, SCO1774, SCO3857, SCO7449 , and hrdB from cDNA prepared from cultures of the parent M145 (marked with W), J2401 (whiA mutant, marked with A) and J2408 (whiH mutant, marked with H) after 18 h, 36 h and 48 h of growth. The assay for each gene was calibrated to the absolute concentration of template per ml reaction volume. Error bars show standard deviations from a total of six assays.
Transcription of SCO1774 and SCO4157 during development of S. coelicolor , analysed by S1 nuclease protection. A. Transcription of SCO1774 in parent strain M145 and J2401 (whiA mutant). B. Transcription of SCO4157 in the parent strain M145, J2401 (whiA mutant) and J2408 (whiH mutant). M marks a lane with a DNA size marker (sizes given in bp). A lane containing a diluted sample of the probe, and another lane with a control reaction with yeast tRNA are indicated. Fragments corresponding to putative transcription start points just upstream of SCO1774 and SCO4157 are indicated by “P”. “R” indicates read-through transcription and “probe” indicates probe-probe reannealing products.
Promoter activity in developing spores. Derivatives of S. coelicolor strain M145 carrying different putative promoters fused to a promoterless mCherry were grown on MS agar to form spores. Spores were analyzed by phase contrast (left panel) and fluorescence microscopy (right panel), to detect the mCherry signal derived from activity of the specific promoters. As controls for hyphal autofluorescence, strain M145 carrying the empty vector pKF210 (A) was also investigated. The investigated putative promoter regions are localized immediately upstream of genes SCO0934 (B), SCO1773 (C), SCO1774 (D), SCO3857 (E), SCO4157 (F), SCO4421 (G), and SCO7449 (H). Representative images are shown here, and quantitative analysis in Table 1. Scale bar, 4μm.
SCO7449-7451– a gene cluster with relation to spore pigmentation
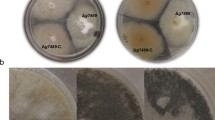
Among the genes showing the largest difference in expression between whi mutants and parent was SCO7449, which encodes a predicted membrane protein of unknown function. The qRT-PCR analysis confirmed the strong up-regulation of SCO7449 during sporulation and showed a strict dependence of this up-regulation on both whiA and whiH (Figure 5). The transcriptional reporter gene construct showed expression specifically in sporulating hyphae (Figure 7). We noted that also the two adjacent genes SCO7450 and SCO7451 (Figure 4) were significantly up-regulated during development of the wild-type strain (Additional file 1: Table S1). These two genes also showed a tendency to be down-regulated in the two whi mutants, although this difference was not statistically significant. We consider it likely that the three genes SCO7449-7451 are co-transcribed. To test whether this group of genes has any function during sporulation, the whole putative operon SCO7449-7451 was deleted and replaced by an apramycin resistance cassette (strain K317). We did not detect any phenotypic effect of the disruption in relation to growth, efficiency of aerial mycelium and spore formation, or shape and stress tolerance of the spores (Figures 8 and 9). However, colonies of the disruption mutant showed a more brownish pigmentation on MS agar compared to the grey appearance of the parent strain, and this change of pigment colour in the mutant could be complemented by the SCO7449-7451 genes integrated at the ϕC31 attB site of the S. coelicolor genome (Figure 8A and C).
Plate phenotypes on MS agar. A. Deletion strains K300 (∆SCO1774-1773), K301 (∆SCO1773), K302 (∆SCO3857), K303 (∆SCO4157), K316 (∆SCO0934), K317 (∆SCO7449-7451), K318 (∆SCO1195-1196), and K319 (∆SCO4421) were grown for three days together with their congenic wild-type parent M145. B. Complementation tests for SCO1774-1773 mutants with cosmid I51, harboring SCO1774-1773 and surrounding sequences. Deletion mutants K300 and K301, wild-type strain M145, and derivatives that had been transformed with cosmid I51, were grown for four days. C. Complementation test for ∆SCO7449-7451 deletion mutant K317 with plasmid pKF278 carrying the SCO7449-7451 locus, and the empty vector pIJ82.
Effect of heat treatment on spores of deletion mutant strains. Spore suspensions of S. coelicolor M145 and the deletion strains K300 (∆SCO1774-1773), K301 (∆SCO1773), K302 (∆SCO3857), K303 (∆SCO4157), K316 (∆SCO0934), K317 (∆SCO7449-7451), K318 (∆SCO1195-1196), and K319 (∆SCO4421) were incubated at 60°C for 30 and 60 minutes. Survival rate of spores was calculated in relation to the number of viable spores in untreated samples. Average values and standard deviations of plate counts from two or three experiments are shown.
SCO7450 encodes a predicted sortase of subgroup E [31], and the SCO7451 gene product shows similarity to proteins associated with polyketide biosynthesis, particularly the S. coelicolor whiE ORFI (SCO5320) product involved in spore pigment biosynthesis, with which it shares 53% identity over 365 amino acids [8]. It has been suggested that whiE ORFI is involved in retaining or targeting the pigment to the spore, possibly within its wall [32]. Comparison of the whiE and SCO7449-7451 regions of the S. coelicolor strain M145 genome to the corresponding sections of three other sequenced streptomycete genomes (S. avermitilis MA-4680, S. clavuligerus ATCC27064, and S. scabies strain 87.22) further supports a link between these two gene clusters and indicates a functional relationship of SCO7451 to spore pigment biosynthesis. The closest homologues of SCO7451 and its two neighboring genes SCO7452 (encoding a putative O-methyltransferase) and SCO7453 (encoding a putative secreted protein) are all found within the whiE gene cluster in the other mentioned genomes, with SCO7451 being most similar to the gene at the position corresponding to whiE ORFI (called sppG in S. avermitilis and S. clavuligerus), and the orthologues of SCO7452 and SCO7453 being located immediately adjacent to the final gene in the spore pigment operon sppE (corresponding to whiE ORFVII). In summary, the altered pigmentation of developing colonies of the ΔSCO7449-7451 mutant, the clear-cut up-regulation of these genes during sporulation, and the linkage of SCO7451 and adjacent genes to orthologues of the whiE gene cluster, lead us to propose an involvement of one or more of the SCO7449-7451 genes in maturation of spores and processing of the spore pigment.
SCO1774-1773– encoding an AfsR-related protein and an L-alanine dehydrogenase
Both genes SCO1773 and SCO1774 showed a whiA-dependent expression according to the microarray data (Figure 2). These genes form a putative transcriptional unit, with SCO1774 encoding a protein with partial similarity to the AfsR regulatory protein [33] and SCO1773 encoding a predicted L-alanine dehydrogenase. The qRT-PCR analyses confirmed the developmental up-regulation of SCO1774 and that this is dependent on whiA (Figure 5). Expression was up-regulated during development of the whiH mutant, but with delay and to a lower level than in the parent strain. The presence of a sporulation-induced promoter for SCO1774, which we here refer to as P1774, was confirmed by the reporter gene assays, which showed high activity in developing spores (Figure 7). S1 nuclease protection assays of SCO1774 identified a putative transcription start site around 30 base pairs upstream of the predicted GTG start codon (Figure 6). This is preceded by an appropriately located -10 promoter motif (TAGGCT), but no corresponding -35 motif could be recognised.
SCO1773 showed a completely different pattern of expression compared to SCO1774, with apparently constitutive presence of the transcript in the wild-type strain, but in agreement with the microarray data, there was a lower level of SCO1773 transcript in the whiA mutant at the 36 and 48 h timepoints compared to the parent strain (Figure 5). To clarify the basis for the differential expression between SCO1774 and SCO1773, the transcripts in this region were investigated using RT-PCR and primer pairs specific to intragenic and intergenic regions of SCO1774 and SCO1773 (Figure 4). Transcripts containing the intragenic region of SCO1773 were abundant, while no transcripts containing the intergenic region between SCO1774 and SCO1773 were detected during vegetative growth (Figure 4 and Additional file 2: Figure S5), suggesting that there is a specific promoter for SCO1773 that is active during vegetative growth. A promoter probe construct carrying parts of the upstream region of SCO1773 failed to detect any activity during vegetative growth or sporulation (Figure 7 and Table 1), but this construct included only 171 base pairs upstream of SCO1773 and the promoter may require additional upstream sequences. During sporulation, transcription from the whiA-dependent P1774 promoter contributes to the expression of SCO1773, as deduced from the presence of transcripts containing the intergenic region between SCO1774 and SCO1773 (Figure 4). This dependence on the P1774 promoter provides a likely explanation of the poor expression of SCO1773 in the whiA mutant (Figures 2 and 5).
Deletion of both SCO1774-1773 (strain K300) or SCO1773 only (strain K301) affected sporulation and resulted in both a reduced spore pigmentation and reduced heat resistance of spores (Figures 8 and 9). A fragment carrying SCO1775-1773 including 240 bp upstream of SCO1775 (Figure 1H) led to partial restoration of the phenotype (data not shown). After complementation with cosmid I51, harboring a larger genomic region around SCO1774-1773, both deletion strains produced the grey spore pigment to the same level as M145 (Figure 8B). It is not clear why the shorter DNA fragments did not lead to full complementation of the mutants. Possibly, even though there is a strongly predicted stem-loop structure immediately after SCO1773 that may serve a transcriptional terminator, polarity on the downstream gene SCO1772 may contribute to the mutant phenotype of the insertions/deletions in SCO1774-1773.
Interestingly, L-alanine dehydrogenase has previously been implicated in development of both Bacillus subtilis and Myxococcus xanthus. Insertions in the ald gene in B. subtilis strongly reduced the efficiency of sporulation [34]. It was speculated that this may be due to a role of alanine dehydrogenase in deaminating the alanine derived from protein turnover and producing pyruvate that can be used for energy metabolism. This was supported by the partial suppression of the ald sporulation phenotype by enriching the medium with pyruvate. The up-regulation of ald transcription during sporulation seemed not to be directly controlled by tested developmental regulators and may be affected by substrate availability or other signals [34]. Mutation of aldA in M. xanthus negatively influenced development, causing delayed aggregation and reduced numbers and viability of spores [35]. The basis for this is unclear, and the required function of alanine dehydrogenase during development appeared not to be production of pyruvate. In similarity to M. xanthus aldA, the SCO1773 mutant phenotype was not affected by enrichment of the medium with pyruvate (data not shown). Nevertheless, the SCO1773 alanine dehydrogenase is required for maturation of spores in S. coelicolor and its expression during sporulation is at least partially achieved by the whiA-dependent promoter P1774.
The SCO1774 gene product shows an interesting similarity to the SARP-type transcription factor AfsR, but it lacks the SARP domain, which is the N-terminal 270 amino acids of AfsR that includes a winged helix motif and a bacterial transcriptional activation domain [33]. Thus, SCO1774 is not likely to encode a transcription factor, and the gene product shows similarity only to the C-terminal parts of AfsR with a tetratricopeptide repeat indicating involvement in protein-protein interactions, and an NB-ARC ATPase domain [36]. In summary, SCO1774 shows a clear-cut developmental transcriptional regulation that is dependent on whiA, but the biological function remains unclear.
SCO3857– encoding a homologue of nosiheptide resistance regulator
SCO3857 encodes a close homologue of the nosiheptide resistance regulatory protein NshA from Streptomyces actuosus with 80.7% identity over the entire sequence of 233 amino acids [37]. Orthologues of SCO3857 are conserved among several streptomycete genomes, including organisms that like S. coelicolor are not resistant to thiopeptide antibiotics like nosiheptide and thiostrepton and do not carry a homologue of the nshR resistance gene that is linked to nshA in S. actuosus. This suggests alternative functions for SCO3857 than control of thiopeptide resistance. The SCO3857 gene showed a clear developmental up-regulation in the wild-type parent, and this was dependent on both whiA and whiH (Figure 5). The mCherry reporter assays showed a high level of expression in sporulating aerial hyphae, but not in vegetative hyphae (Figure 7). Finally, although a SCO3857 deletion mutant produced normal-looking colonies on MS agar (Figure 8), we detected a reduced heat-resistance of the mutant spores compared to the parent strain (Figure 9). These observations identify SCO3857 as a sporulation gene with a role in maturation of spores.
Other developmentally regulated loci
The SCO4421 gene encodes a TetR family regulator and is located close to afsK (SCO4423), which encodes a Ser/Thr protein kinase involved in apical growth and branching of hyphae, as well as in control of secondary metabolism [38, 39]. SCO4421 showed statistically significant up-regulation in the parent strain M145 and decreased expression in the whiA mutant in the array data (Figure 2 and Additional file 1: Table S1). The developmental regulation was not tested by qRT-PCR, but was confirmed by the mCherry reporter construct that showed clear signal in spore chains but not in vegetative hyphae (Figure 7 and Table 1). We did not detect any phenotype associated with the SCO4421 deletion mutant (Figure 8), and its function during sporulation therefore remains unclear.
SCO4157 encodes a putative trypsin-like serine protease. The developmental up-regulation and the decreased expression in both whiA and whiH mutants was confirmed by S1 nuclease protection assays (Figure 6B). The assays pinpointed a 5′-end for SCO4157 transcripts that overlaps with the predicted translational start, and this signal was strongly increased during development of strain M145, but was much weaker in the whiA mutant. A delayed up-regulation was seen in the whiH strain (Figure 6B). Further, there is contribution from promoters located upstream of the probe used in these assays, possibly from the SCO4158 gene. The mCherry reporter gene assays for SCO4157 showed a low but significant signal in developing spores (Figure 7 and Table 1), further supporting that SCO4157 is expressed during sporulation. The discovery of a protease that is expressed during sporulation is interesting in relation to the known involvement of extracellular proteases and protease inhibitors in controlling development of S. coelicolor and other streptomycetes [3, 40]. However, no phenotype was detected in the SCO4157 deletion mutant, and the absence of unequivocal secretion signals in the amino acid sequence makes the role of the SCO4157 protease in such extracellular signalling unclear.
The microarray analyses showed significant changes of expression for SCO0934, with decreased levels of transcripts in both mutants (Figure 2 and Additional file 1: Table S1). The developmental up-regulation in the wild-type strain and the lower transcript levels in the mutants were confirmed by qRT-PCR, although there was a limited up-regulation of this gene in the whi mutants. A low but significant signal was detected in spores from the SCO0934 promoter probe construct, but no phenotype was revealed in the SCO0934 deletion mutant (Figure 7 and Table 1). Thus, it remains unclear whether there is a sporulation-related role for this gene, which encodes a predicted membrane protein of unknown function.
SCO1195 encodes a small predicted membrane protein with similarity to the previously described SmeA protein that is produced during sporulation of S. coelicolor[41]. SmeA is required for the targeting of SffA, a protein with similarity to the SpoIIIE/FtsK family of DNA transporters, to sporulation septa, and several of the SmeA homologues in streptomycetes are encoded together with members of this protein family [41]. This is not the case for SCO1195, which instead may be co-transcribed with SCO1196, encoding a known substrate for secretion via the Tat pathway but of unknown function [42]. The results on SCO1195 expression were similar to those of SCO0934, with significant developmental up-regulation in the parent strain, lower expression in the whiA strain detected in the array experiments (Figure 2), and confirmation of this by real-time qRT-PCR (Figure 5). A SCO1195-1196 deletion mutant failed to reveal any obvious phenotype.
Conclusions
The aerial hyphal sporulation in S. coelicolor occurs only in a fraction of the colony biomass and is not highly synchronized. Thus, even if a gene is strongly induced at a specific stage of sporulation, it is highly challenging to detect this change in global transcriptome investigations of total RNA extracted from the complex mixtures of cell-types that constitute a developing Streptomyces colony. We show here that by comparing a wild-type to mutants lacking key regulators that specifically act in processes linked to aerial hypha, it is possible to identify previously unknown genes that are up-regulated in sporulating aerial hyphae. These genes are not necessarily direct targets for transcriptional regulation by the WhiA or WhiH proteins. In fact, there is no clear ovelap between the set of genes identified here and the very recently described direct targets of WhiA in Streptomyces venezuelae[43]. Nevertheless, our approach allowed identification of several new genes that are important for sporulation in S. coelicolor. Some of the developmentally regulated genes that were found by this transcriptome analysis have been investigated here and in a previous study [30], and the function and regulation of others remain now to be investigated in detail.
Methods
Strains and growth conditions
Bacterial strains used are shown in Table 2. E. coli strain DH5α was used as a host for plasmid construction and strain ET12567/pUZ8002 was used to drive conjugative transfer of nonmethylated plasmid DNA to S. coelicolor A3(2) strains, which have a methyl-specific restriction system. E. coli strain DY380 was used for λRED-mediated recombination to replace target S. coelicolor genes on cosmids with antibiotic resistance cassettes [44]. S. coelicolor A3(2) strain M145 and its derivates were grown at 30°C on Mannitol Soya flour (MS) agar or in yeast extract malt extract (YEME) medium [45]. Media used for E. coli strains were Difco nutrient agar and broth if viomycin was used for selection and Luria-Bertani media for other antibiotics. Antibiotics were used at the following concentrations: apramycin 25 μg ml-1, nalidixic acid 20 μg ml-1, viomycin 30 μg ml-1, and kanamycin 5 μg ml-1 for S. coelicolor, and carbenicillin 100 μg ml-1, kanamycin 50 μg ml-1, viomycin 30 μg ml-1, and apramycin 50 μg ml-1 for E. coli.
General molecular techniques
General DNA manipulations and cloning were carried out as described previously [30]. The oligonucleotide primers used in this study are listed in Additional file 3: Table S2.
Preparation of total RNA from S. coelicolorstrains for microarray experiment, RT-PCR, and S1 nuclease protection assays
S. coelicolor M145 and non-sporulating strains J2408 (ΔwhiH::ermE) and J2401 (whiA::hyg) were pre-cultivated in 25 ml of YEME medium. M145 was grown for 20–22 h and the whi mutants J2408 and J2401 for 40–44 h to reach similar cell densities. The mycelium was harvested and washed twice with water, ground in 3 ml 10.3% (w/v) sucrose in a glass homogenizer and sonicated for 10 min in a sonic bath to disrupt clumped mycelia. This allowed the inoculation of plates for the array analyses in an equivalent way for both sporulating and non-sporulating strains. About 3x106 colony forming units were inoculated onto cellophane-coated MS plates to obtain a confluent growth. Mycelium was scraped from the cellophane discs at three different times during development: 18 h when only bald vegetative mycelium was observed, 36 h when thin aerial mycelium was covering the plates, and 48 h when mycelium surface was grey due to abundant sporulation. Cells were harvested from 2 to 12 plates to get ca. 30 mg of dry weight cells per time point and strain. Harvested mycelia were treated with RNAprotect Bacteria Reagent (Qiagen) to stabilize the RNA. Cell lysis, RNA isolation and DNAase treatment were then carried out using the Total RNA Isolation with RNeasy Protect Bacteria Kit described in http://www2.surrey.ac.uk/fhms/microarrays/. The RNA samples were subjected to quality control by the Bioanalyzer RNA 6000 Nano Assay (Agilent Technologies) and only RNA with an RNA integrity number (RIN) between 7 and 10 were taken forward to further analysis (microarray experiments, qRT-PCR, S1 nuclease protection assay, and the reverse transcription PCR).
Microarray experiments and data analysis
Total RNA was isolated at three time points for each strain from four replicated cultures. RNA samples of the four biological replicates were reverse-transcribed and labeled according to the protocols detailed in http://www2.surrey.ac.uk/fhms/microarrays/Downloads/Protocols/. For each time-point and strain the cDNA samples from two biological replicates were labeled with Cy3 and two with Cy5. Each mutant cDNA sample was cohybridised with the corresponding (matched timepoints and opposite dye orientation) wild-type cDNA to arrays according to a ‘Balanced Block Design’ [27], as outlined in Figure 1. In addition, direct comparisons of M145 48 h vs M145 18 h and M145 36 h vs M145 18 h cDNA were conducted, also with a balanced block design, to reveal genes changing during normal development of the wild type. Thus, a total of 32 arrays were used in this analysis.
After scanning with an Affymetrix 428 array scanner, the images were processed with BlueFuse 3.1 software (BlueGnome). Array data were analyzed using R [54] and the Bioconductor [55] package limma [56, 57]. Raw data were transformed to log2 scale and normalized by applying print-tip loess to each array followed by an across array normalisation (‘scale’ function in the limma package). Because equal dyes are needed in the balanced block design, only genes having at least one good spot on all four arrays of a particular comparison were considered in further analysis. Differential significance between conditions was determined by using the eBayes function of limma; resultant p-values were corrected by the application of Benjamini and Hochberg “false discovery rate” correction [28]. A difference in gene expression was considered significant if it had an adjusted p-value <0.05. The microarray data have been deposited with ArrayExpress (Accession number E-MTAB-1942).
Quantitative real time PCR (qRT-PCR)
RNA samples, isolated as described above, were further treated with RQ1 RNase-free DNase (Promega) to remove all traces of DNA. DyNAmo™ SYBR® Green 2-Step qRT-PCR kit (Finnzymes) was used to generate cDNA and reactions were carried out at 45°C for 1 h using 15 ng of random hexamers primers and 1 μg of total RNA. Two biological replicates of the RNA were used and three independent qRT-PCR reactions were run for each of them, i.e. six in total for each strain and time point. Quantitative real-time PCR of selected genes was performed using a Rotor-Gene 2000 Real-time cycler (Corbett Research). Two μl of a 1:5 dilution (in 10 mM Tris–HCl pH 8.0) of first strand cDNA reaction was used as a DNA template in a 20 μl final reaction volume of the qPCR using a specific primer pair for each tested gene (Additional file 3: Table S2). hrdB is a constitutively expressed gene encoding the principal RNA polymerase factor of S. coelicolor, and was used as a control for the qRT-PCR experiment. Negative controls with 10 mM Tris–HCl pH 8.0 instead of template were included. To quantitate the abundance of a specific transcript, standard curves were generated using appropriate dilutions of a DNA template of known concentration for each one of the tested genes, and the averaged copy number of six independent q-RT-PCR reactions, calculated in relation to the standard curve, was calculated.
S1-nuclease mapping
For each S1 nuclease reaction, 30 μg of total RNA, prepared as described above, was hybridized to a radioactive probe prepared by PCR. First, a region spanning the presumed promoter region upstream of the first start codon was amplified using primers KF260 and KF261 for SCO1774 and KF256 and KF257 for SCO4157 (Additional file 3: Table S2). The resulting PCR products were cloned in pCR-BluntII TOPO vector. The reverse primers (KF261, and KF257) were phosphorylated using γ-32P ATP before use in amplification. Together with a forward primer in the vector sequence, it generated a PCR fragment uniquely labeled on the reverse strand and containing a non-homologous upstream extension (about 150 nucleotides) to discriminate between full-length protection and probe-probe re-annealing products. S1 nuclease protection was carried out as described previously [58]. Approximately 30.000 Cerenkov count min-1 of the labeled probe was used in each hybridization reaction. S1 digestion (Fermentas S1 nuclease) was performed for 1 h at 37°C and digestion products were separated on an 8% denaturing polyacrylamide gel. Molecular weight markers were produced by end-labeling of MspI-digested pBR322.
Reverse transcription assay of transcripts from the SCO1774-1773locus
cDNA, prepared as described above from RNA isolated from strain M145 after 18 h and 48 h, was used as a template in PCR amplifications. Different primer pairs (Additional file 3: Table S2) were used to detect the presence of transcripts; primers 4-3for and 4-3rev to detect transcripts spanning the intergenic regions between SCO1774 and SCO1773; 1774RTfor and 1774RTrev to detect transcripts including intragenic regions of SCO1774; and 1773RTfor and 1773RTrev to detect transcripts including intragenic regions of SCO1773. A control without reverse transcriptase was included to confirm that detected products did not derive from amplification of contaminating DNA in the RNA preparations, and a positive control that used genomic DNA as template was also included.
Construction of S. coelicolordisruption mutants
For generation of gene deletion mutants in S. coelicolor strain M145, λRED-mediated PCR-targeting was carried out as described previously [59]. The primers used to amplify the disruption cassettes are listed in Additional file 3: Table S2. They were amplified from pIJ773 containing the apramycin resistance gene aac(3)IV, pIJ780 containing the viomycin resistance gene vph, and plasmid pHP45Ωaac containing the apramycin resistance cassette ΩaacC4. The targeted genes were first disrupted on cosmids (listed in Table 2) in E. coli strain DY380. Mutated cosmids were introduced into S. coelicolor by protoplast transformation (for mutant alleles constructed with ΩaacC4 since this does not carry an oriT) or conjugation (for pIJ773 and pIJ780-derived cassettes that carry oriT), and clones were identified in which a double cross-over event had led to replacement of the target gene with the disruption allele present on the cosmids. The gene replacements were confirmed with Southern blotting and PCR (data not shown).
Complementation constructs
The disruption mutants K300 (ΔSCO1774-1773::vph) and K301 (ΔSCO1773::vph) were tested for complementation using a 4.6 kb fragment containing SCO1775-SCO1773 coding regions, including 240 bp upstream of the SCO1775 and 343 bp downstream of SCO1773. This fragment was amplified from cosmid I51 using primers KF487 and KF488 and cloned in a pCR-BluntII vector. The cloned fragment was cut out using XbaI and HindIII restriction sites in the vector and ligated into pOJ260 cut with the same enzymes.
Complementation of deletion strain K317 (ΔSCO7449-7451::aac(3)IV) was carried out using a 3.5 kb fragment that included all three genes and 487 bp upstream of SCO7449 and 245 bp downstream of SCO7451. This was amplified from cosmid 5C11 using primer KF527 and KF528, cloned in the pCR-BluntII vector, recovered using BamHI and XbaI restriction sites in the vector, and cloned in pIJ82 for transfer to the S. coelicolor strains.
Construction of promoter fusions to the mCherry reporter gene
The promoter-probe vector pKF210 was designed to facilitate construction of promoter fusions to the gene for mCherry fluorescent protein. Most of the vector pIJ6902, except the inducible tipA promoter, was amplified by PCR with phosphorylated primers TL03 (adding an EcoRI site) and TL04 (adding a NotI site). The gene encoding mCherry was amplified from pKS-mCherry-S-T3 using primer TL01, containing an EcoRI site followed by BamHI and XbaI sites, a ribosome binding site, and finally an NdeI site overlapping the start codon of the mCherry coding region, and primer TL02, which included a NotI site. The two PCR products were digested with EcoRI and NotI and ligated to form pKF210.
The promoter regions of SCO0934 (including a 203 bp segment upstream from the start codon and the first14 codons of the gene), SCO1773 (including 171 bp upstream of the start codon and 16 codons of the gene), SCO1774 (including 273 bp upstream of the start codon and 13 codons of the gene), SCO3857 (including 368 bp upstream of the start codon and 17 codons of the gene), SCO4157 (including 152 bp upstream of the start codon and 14 codons of the gene), SCO4421 (including 170 bp of the upstream region and 22 codons from the gene) and SCO7449 (including 282 bp of the upstream region and 11 codons from the gene) were amplified using forward and a reverse primers with 5′-tails containing XbaI and NdeI sites (Additional file 3: Table S2), and ligated into pKF210 to make translational fusions to mCherry.
Microscopy
For detection and quantification of the mCherry signal, strains were grown in liquid culture in tryptic soy broth (TSB) to obtain growing vegetative mycelium, and on MS agar for spore formation. Vegetative hyphae were added directly to slides coated with 1% (w/v) agarose in phosphate-buffered saline. Spore chains were collected by pressing coverslips on the surface of colonies and then placing them on agarose-coated slides. Images of fluorescence signals were captured and analysed quantitatively using a previously described microcopy system [30]. Aerial mycelium and spores of all mutants were also investigated by phase-contrast microscopy.
Heat resistance of spores
The ability of spores to survive incubation at 60°C was assayed as described previously [30].
Availability of supporting data
The microarray data has been deposited with ArrayExpress (Accession number: E-MTAB-1942).
References
Chater KF: Differentiation in Streptomyces: the properties and programming of diverse cell-types. Streptomyces: Molecular Biology and Biotechnology. Edited by: Dyson P. 2011, Norfolk, UK: Caister Academic Press, 43-86.
Flärdh K, Buttner MJ: Streptomyces morphogenetics: Dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol. 2009, 7: 36-49. 10.1038/nrmicro1968.
Chater KF, Biro S, Lee KJ, Palmer T, Schrempf H: The complex extracellular biology of Streptomyces. FEMS Microbiol Rev. 2010, 34 (2): 171-198. 10.1111/j.1574-6976.2009.00206.x.
McCormick JR, Flärdh K: Signals and regulators that govern Streptomyces development. FEMS Microbiol Rev. 2012, 36 (1): 206-231. 10.1111/j.1574-6976.2011.00317.x.
Van Wezel GP, McDowall KJ: The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep. 2011, 28 (7): 1311-1333. 10.1039/c1np00003a.
Bibb MJ, Domonkos A, Chandra G, Buttner MJ: Expression of the chaplin and rodlin hydrophobic sheath proteins in Streptomyces venezuelae is controlled by sigma(BldN) and a cognate anti-sigma factor, RsbN. Mol Microbiol. 2012, 84 (6): 1033-1049. 10.1111/j.1365-2958.2012.08070.x.
Den Hengst CD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, Buttner MJ: Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol Microbiol. 2010, 78 (2): 361-379. 10.1111/j.1365-2958.2010.07338.x.
Davis NK, Chater KF: Spore colour in Streptomyces coelicolor A3(2) involves the developmentally regulated synthesis of a compound biosynthetically related to polyketide antibiotics. Mol Microbiol. 1990, 4: 1679-1691. 10.1111/j.1365-2958.1990.tb00545.x.
Kelemen GH, Brian P, Flärdh K, Chamberlin LC, Chater KF, Buttner MJ: Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J Bacteriol. 1998, 180 (9): 2515-2521.
Chater KF, Bruton CJ, Plaskitt KA, Buttner MJ, Méndez C, Helmann JD: The developmental fate of S. coelicolor hyphae depends on a gene product homologous with the motility σ factor of B. subtilis. Cell. 1989, 59: 133-143. 10.1016/0092-8674(89)90876-3.
Kelemen GH, Brown GL, Kormanec J, Potúcková L, Chater KF, Buttner MJ: The positions of the sigma factor genes whiG and sigF in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolor A3(2). Mol Microbiol. 1996, 21 (3): 593-603. 10.1111/j.1365-2958.1996.tb02567.x.
Aínsa JA, Parry HD, Chater KF: A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2). Mol Microbiol. 1999, 34 (3): 607-619. 10.1046/j.1365-2958.1999.01630.x.
Ryding NJ, Kelemen GH, Whatling CA, Flärdh K, Buttner MJ, Chater KF: A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2). Mol Microbiol. 1998, 29 (1): 343-357. 10.1046/j.1365-2958.1998.00939.x.
Chater KF: Construction and phenotypes of double sporulation deficient mutants in Streptomyces coelicolor A3(2). J Gen Microbiol. 1975, 87: 312-325. 10.1099/00221287-87-2-312.
Flärdh K, Findlay KC, Chater KF: Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2). Microbiology. 1999, 145 (Pt 9): 2229-2243.
Persson J, Chater KF, Flärdh K: Molecular and cytological analysis of the expression of Streptomyces sporulation regulatory gene whiH. FEMS Microbiol Lett. 2013, 341 (2): 96-105. 10.1111/1574-6968.12099.
Tian Y, Fowler K, Findlay K, Tan H, Chater KF: An unusual response regulator influences sporulation at early and late stages in Streptomyces coelicolor. J Bacteriol. 2007, 189 (7): 2873-2885. 10.1128/JB.01615-06.
Zhang G, Tian Y, Hu K, Zhu Y, Chater KF, Feng C, Liu G, Tan H: Importance and regulation of inositol biosynthesis during growth and differentiation of Streptomyces. Mol Microbiol. 2012, 83 (6): 1178-1194. 10.1111/j.1365-2958.2012.08000.x.
Aínsa JA, Ryding NJ, Hartley N, Findlay KC, Bruton CJ, Chater KF: WhiA, a protein of unknown function conserved among Gram-positive bacteria, is essential for sporulation in Streptomyces coelicolor A3(2). J Bacteriol. 2000, 182 (19): 5470-5478. 10.1128/JB.182.19.5470-5478.2000.
Kaiser BK, Clifton MC, Shen BW, Stoddard BL: The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease. Structure. 2009, 17 (10): 1368-1376. 10.1016/j.str.2009.08.008.
Knizewski L, Ginalski K: Bacterial DUF199/COG1481 proteins including sporulation regulator WhiA are distant homologs of LAGLIDADG homing endonucleases that retained only DNA binding. Cell Cycle. 2007, 6 (13): 1666-1670. 10.4161/cc.6.13.4471.
Kaiser BK, Stoddard BL: DNA recognition and transcriptional regulation by the WhiA sporulation factor. Sci Rep. 2011, 1: 156-
Davis NK, Chater KF: The Streptomyces coelicolor whiB gene encodes a small transcription factor-like protein dispensable for growth but essential for sporulation. Mol Gen Genet. 1992, 232: 351-358.
Soliveri JA, Gomez J, Bishai WR, Chater KF: Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology. 2000, 146 (Pt 2): 333-343.
Crack JC, Den Hengst CD, Jakimowicz P, Subramanian S, Johnson MK, Buttner MJ, Thomson AJ, Le Brun NE: Characterization of [4Fe-4S]-containing and cluster-free forms of Streptomyces WhiD. Biochemistry. 2009, 48 (51): 12252-12264. 10.1021/bi901498v.
Facey PD, Sevcikova B, Novakova R, Hitchings MD, Crack JC, Kormanec J, Dyson PJ, Del Sol R: The dpsA gene of Streptomyces coelicolor: Induction of expression from a single promoter in response to environmental stress or during development. PLoS One. 2011, 6 (9): e25593-10.1371/journal.pone.0025593.
Dobbin K, Shih JH, Simon R: Statistical design of reverse dye microarrays. Bioinformatics. 2003, 19 (7): 803-810. 10.1093/bioinformatics/btg076.
Benjamini Y, Hochberg Y: Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Statist Soc B. 1995, 57 (1): 289-300.
Saito N, Xu J, Hosaka T, Okamoto S, Aoki H, Bibb MJ, Ochi K: EshA accentuates ppGpp accumulation and is conditionally required for antibiotic production in Streptomyces coelicolor A3(2). J Bacteriol. 2006, 188 (13): 4952-4961. 10.1128/JB.00343-06.
Salerno P, Larsson J, Bucca G, Laing E, Smith CP, Flärdh K: One of the two genes encoding nucleoid-associated HU proteins in Streptomyces coelicolor is developmentally regulated and specifically involved in spore maturation. J Bacteriol. 2009, 191 (2): 6489-6500.
Marraffini LA, Dedent AC, Schneewind O: Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006, 70 (1): 192-221. 10.1128/MMBR.70.1.192-221.2006.
Yu T-W, Hopwood DA: Ectopic expression of the Streptomyces coelicolor whiE genes for polyketide spore pigment synthesis and their interaction with the act genes for actinorhodin biosynthesis. Microbiology. 1995, 141: 2779-2791. 10.1099/13500872-141-11-2779.
Tanaka A, Takano Y, Ohnishi Y, Horinouchi S: AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J Mol Biol. 2007, 369 (2): 322-333. 10.1016/j.jmb.2007.02.096.
Siranosian KJ, Ireton K, Grossman AD: Alanine dehydrogenase (ald) is required for normal sporulation in Bacillus subtilis. J Bacteriol. 1993, 175 (21): 6789-6796.
Ward MJ, Lew H, Zusman DR: Disruption of aldA influences the developmental process in Myxococcus xanthus. J Bacteriol. 2000, 182 (2): 546-550. 10.1128/JB.182.2.546-550.2000.
van der Biezen EA, Jones JD: The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol. 1998, 8 (7): R226-R227. 10.1016/S0960-9822(98)70145-9.
Li Y, Dosch DC, Woodman RH, Floss HG, Strohl WR: Transcriptional organization and regulation of the nosiheptide resistance gene in Streptomyces actuosus. J Ind Microbiol. 1991, 8 (1): 1-12. 10.1007/BF01575585.
Hempel AM, Cantlay S, Molle V, Wang SB, Naldrett MJ, Parker JL, Richards DM, Jung YG, Buttner MJ, Flärdh K: The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proc Natl Acad Sci USA. 2012, 109 (35): E2371-E2379. 10.1073/pnas.1207409109.
Umeyama T, Lee P-C, Horinouchi S: Protein serine/threonine kinases in signal transduction for secondary metabolism and morphogenesis in Streptomyces. Appl Microbiol Biotechnol. 2002, 59: 419-425. 10.1007/s00253-002-1045-1.
Kim DW, Hesketh A, Kim ES, Song JY, Lee DH, Kim IS, Chater KF, Lee KJ: Complex extracellular interactions of proteases and a protease inhibitor influence multicellular development of Streptomyces coelicolor. Mol Microbiol. 2008, 70 (5): 1180-1193. 10.1111/j.1365-2958.2008.06471.x.
Ausmees N, Wahlstedt H, Bagchi S, Elliot MA, Buttner MJ, Flärdh K: SmeA, a small membrane protein with multiple functions in Streptomyces sporulation including targeting of a SpoIIIE/FtsK-like protein to cell division septa. Mol Microbiol. 2007, 65 (6): 1458-1473. 10.1111/j.1365-2958.2007.05877.x.
Widdick DA, Dilks K, Chandra G, Bottrill A, Naldrett M, Pohlschroder M, Palmer T: The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proc Natl Acad Sci USA. 2006, 103 (47): 17927-17932. 10.1073/pnas.0607025103.
Bush MJ, Bibb MJ, Chandra G, Findlay KC, Buttner MJ: Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae. MBio. 2013, 4 (5): e00684-00613-
Yu D, Ellis HM, Lee E-C, Jenkins NA, Copeland NG, Court DL: An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000, 97: 5978-5983. 10.1073/pnas.100127597.
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA: Practical Streptomyces Genetics. 2000, The John Innes Foundation: Norwich, UK
Lee E-C, Yu D, DVJ M, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG: A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001, 73: 56-65. 10.1006/geno.2000.6451.
Gust B, Challis GL, Fowler K, Kieser T, Chater KF: PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003, 100 (4): 1541-1546. 10.1073/pnas.0337542100.
Blondelet-Rouault M-H, Weiser J, Lebrihi A, Branny P, Pernodet J-L: Antibiotic resistance gene cassettes derived from the ω interposon for use in E. coli and Streptomyces. Gene. 1997, 190: 315-317. 10.1016/S0378-1119(97)00014-0.
Janssen GR, Bibb MJ: Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene. 1993, 124 (1): 133-134. 10.1016/0378-1119(93)90774-W.
Bierman M, Logan R, O’Brien K, Seno ET, Rao RN, Schoner BE: Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992, 116 (1): 43-49. 10.1016/0378-1119(92)90627-2.
Gregory MA, Till R, Smith MC: Integration site for Streptomyces phage phiBT1 and development of site-specific integrating vectors. J Bacteriol. 2003, 185 (17): 5320-5323. 10.1128/JB.185.17.5320-5323.2003.
Huang J, Lih CJ, Pan KH, Cohen SN: Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 2001, 15 (23): 3183-3192. 10.1101/gad.943401.
Redenbach M, Kieser HM, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood DA: A set of ordered cosmids and a detailed genetic and physical map of the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996, 21 (1): 77-96. 10.1046/j.1365-2958.1996.6191336.x.
R: A language and environment for statistical computing. http://www.R-project.org,
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al: Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004, 5 (10): R80-10.1186/gb-2004-5-10-r80.
Smyth GK: Limma: linear models for microarray data. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Edited by: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W. 2005, New York: Springer, 397-420.
Smyth GK, Speed TP: Normalization of cDNA microarray data. Methods. 2003, 31: 265-273. 10.1016/S1046-2023(03)00155-5.
Flärdh K, Leibovitz E, Buttner MJ, Chater KF: Generation of a non-sporulating strain of Streptomyces coelicolor A3(2) by the manipulation of a developmentally controlled ftsZ promoter. Mol Microbiol. 2000, 38 (4): 737-749. 10.1046/j.1365-2958.2000.02177.x.
Flärdh K: Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol Microbiol. 2003, 49 (6): 1523-1536. 10.1046/j.1365-2958.2003.03660.x.
Acknowledgements
This work was supported by postdoctoral stipends from Carl Tryggers Foundation to PS and NA, and by grants from the Swedish Research Council (No. 621-2007-4767) to KF and the European Commission FP6 Programme,(No, IP005224, ActinoGEN) to CPS.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
PS prepared all biological material for the array experiment, and carried out the array hybridizations and data analyses together with GB, EL, and CPS, who contributed materials, technology and knowhow for the transcriptome experiments. EL contributed particularly to the bioinformatic analyses. PS also carried out the qRT-PCR and S1 nuclease protection assays. JP, PS, and NA constructed the relevant mutants, and JP analysed phenotypes, and carried out the fluorescence-based promoter-probe experiments. KF, PS, and JP planned the work, and KF and JP wrote the paper, with contributions from all of the other authors. All authors read and approved the final manuscript.
Electronic supplementary material
12866_2013_2163_MOESM1_ESM.xlsx
Additional file 1: Table S1: Genes that are differentially expressed when comparing whiA or whiH mutant to the wild-type parent, or comparing the developing wild-type strain at 36 h or 48 h to the expression pattern at 18 h. All ORFs having an adjusted p-value <0.05 in at least one of the eight comparisons (A18, A36, A48, H18, H36, H48, wt36, wt 48) are listed. There are 285 ORFs in total. (XLSX 47 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Salerno, P., Persson, J., Bucca, G. et al. Identification of new developmentally regulated genes involved in Streptomyces coelicolorsporulation. BMC Microbiol 13, 281 (2013). https://doi.org/10.1186/1471-2180-13-281
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-13-281