Abstract
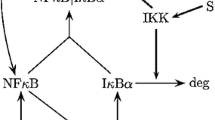
The regulation of I B, NF-
B, NF- B is of foremost interest in biology as the transcription factor NF-
B is of foremost interest in biology as the transcription factor NF- B has multiple target genes. We have modeled a previously published model by Hoffmann et al. (2002) of I
B has multiple target genes. We have modeled a previously published model by Hoffmann et al. (2002) of I B, NF-
B, NF- B mathematically as discrete reaction systems. We have used stochastic algorithm to compare the results when there are large and small numbers of molecules available in a finite volume for each protein. Our results for small number of molecules show that with continuous presence of stimulation, nuclear NF-
B mathematically as discrete reaction systems. We have used stochastic algorithm to compare the results when there are large and small numbers of molecules available in a finite volume for each protein. Our results for small number of molecules show that with continuous presence of stimulation, nuclear NF- B oscillates continuously in every individual cell rather than damping, which was observed in cell population results. This characteristic of the system is missed when averaged behavior is studied.
B oscillates continuously in every individual cell rather than damping, which was observed in cell population results. This characteristic of the system is missed when averaged behavior is studied.
Similar content being viewed by others
References
Ghosh S, May MJ, Kopp EB:NF-
 B and rel proteins: evolutionary conserved mediators of immune responses. Annual Review of Immunology 1998, 16: 225-260. 10.1146/annurev.immunol.16.1.225
B and rel proteins: evolutionary conserved mediators of immune responses. Annual Review of Immunology 1998, 16: 225-260. 10.1146/annurev.immunol.16.1.225Gonzalez-Crespo S, Levine M:Related target enhancers for dorsal and NF-
 B signaling pathways. Science 1994, 264(5156):255-258. 10.1126/science.8146656
B signaling pathways. Science 1994, 264(5156):255-258. 10.1126/science.8146656Malek S, Huxford T, Ghosh G:I
 B
B functions through direct contacts with the nuclear localization signals and the DNA binding sequences of NF-
functions through direct contacts with the nuclear localization signals and the DNA binding sequences of NF- B. Journal of Biological Chemistry 1998, 273(39):25427-25435. 10.1074/jbc.273.39.25427
B. Journal of Biological Chemistry 1998, 273(39):25427-25435. 10.1074/jbc.273.39.25427Hoffmann A, Levchenko A, Scott ML, Baltimore D:The I
 B-NF-
B-NF- B signaling module: temporal control and selective gene activation. Science 2002, 298(5596):1241-1245. 10.1126/science.1071914
B signaling module: temporal control and selective gene activation. Science 2002, 298(5596):1241-1245. 10.1126/science.1071914Nelson DE, Ihewaba AEC, Elliott M, et al.:Oscillations in NF-
 B signaling control the dynamics of gene expresion. Science 2004, 306(5696):704-708. 10.1126/science.1099962
B signaling control the dynamics of gene expresion. Science 2004, 306(5696):704-708. 10.1126/science.1099962Barken D, Wang CJ, Kearns J, Cheong R, Hoffmann A, Levchenko A:Comment on oscillations in NF-
 B signaling control the dynamics of the gene expression. Science 2005, 308(5718):52. 10.1126/science.1107904
B signaling control the dynamics of the gene expression. Science 2005, 308(5718):52. 10.1126/science.1107904Nelson DE, Horton CA, See V, et al.:Response to comment on oscillations in NF-
 B signaling control the dynamics of the gene expression. Science 2005, 308(5718):52. 10.1126/science.1107904
B signaling control the dynamics of the gene expression. Science 2005, 308(5718):52. 10.1126/science.1107904Lauffenburger DA: Receptors: Models for Binding, Trafficking and Signaling. Oxford University Press, Oxford, UK; 1993.
Bhalla US, Iyengar R: Emergent properties of networks of biological signaling pathways. Science 1999, 283(5400):381-387. 10.1126/science.283.5400.381
Kholodenko BN: Negative feedbacka dn ultrasensitivity can bring about oscillationsin the mitogen-activated protein kinase cascades. European Journal of Biochemistry 2000, 267(6):1583-1588.
Bentele M, Lavrik I, Ulrich M, et al.: Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis. Journal of Cell Biology 2004, 166(6):839-851. 10.1083/jcb.200404158
Luby-Phelps K, Weisiger RA: Role of cytoarchitecture in cytoplasmic transport. Comparative Biochemistry and Physiology B 1996, 115(3):295-306. 10.1016/S0305-0491(96)00176-9
Goodsell DS: Inside a living cell. Trends in Biochemical Sciences 1991, 16(6):203-206.
Berg OG: The influence of macromolecular crowding on thermodynamic activity: solubility and dimerization constants for spherical and dumbbell-shaped molecules in a hard-sphere mixture. Biopolymers 1990, 30(11-12):1027-1037. 10.1002/bip.360301104
Han J, Herzfeld J: Macromolecular diffusion in crowded solutions. Biophysical Journal 1993, 65(3):1155-1161. 10.1016/S0006-3495(93)81145-7
Lahav G, Rosenfeld N, Sigal A, et al.: Dynamcis of the p53-Mdm2 feedback loop in individual cells. Nature Genetics 2004, 36(2):147-150. 10.1038/ng1293
Vilar JMG, Kueh HY, Barkai N, Leibler S: Mechanisms of noise-resistance in genetic oscillators. Proceedings of the National Academy of Sciences of the United States of America 2002, 99(9):5988-5992. 10.1073/pnas.092133899
Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP: Causal protein-signaling networks derived from multiparameter single-cell data. Science 2005, 308(5721):523-529. 10.1126/science.1105809
Mcadams HH, Arkin A: Stochastic mechanisms in gene expression. Proceedings of the National Academy of Sciences of the United States of America 1997, 94(3):814-819. 10.1073/pnas.94.3.814
McAdams H, Arkin H: It's noisy business! Genetic regulation at the nanomolecular scale. Trends Genetics 1999, 15(2):65-69. 10.1016/S0168-9525(98)01659-X
Gonze D, Halloy J, Goldbeter A: Robustness of circadian rhythms with respect to molecular noise. Proceedings of the National Academy of Sciences of the United States of America 2002, 99(2):673-678. 10.1073/pnas.022628299
Srivastava R, You L, Summers J, Yin J: Stochastic vs deterministic modeling of intracellular viral kinetics. Journal of Theoretical Biology 2002, 218(3):309-321. 10.1006/jtbi.2002.3078
Bhalla US: Signaling in small subcellular volumes. I. Stochastic and diffusion effects on individual pathways. Biophysical Journal 2004, 87(2):733-744. 10.1529/biophysj.104.040469
Bhalla US: Signaling in small subcellular volumes. II. Stochastic and diffusion effects on synaptic network properties. Biophysical Journal 2004, 87(2):745-753. 10.1529/biophysj.104.040501
[http://www.cellsystems.org/teams/modeling/projects/sigtran/index.html]
Morton-Firth CJ, Bray D: Predicting temporal fluctuations in an intracellular signalling pathway. Journal of Theoretical Biology 1998, 192(1):117-128. 10.1006/jtbi.1997.0651
Gillepsie DT: A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. Journal of Computational Physics 1976, 22(4):403-434. 10.1016/0021-9991(76)90041-3
Gibson MA, Bruck J: Efficient exact stochastic simulation of chemical systems with many species and many channels. Journal of Physical Chemistry A 2000, 104(9):1876-1889. 10.1021/jp993732q
Gillespie DT: Exact stochastic simulation of coupled chemical reactions. Journal of Physical Chemistry 1977, 81(25):2340-2361. 10.1021/j100540a008
Elliott DF: Handbook of Digital Signal Processing: Engineering Applications. Academic Press, New York, NY, USA; 1987.
Grenander U, Szego G: Toeplitz Forms and Their Applications. Chelsea, New York, NY, USA; 1984.
Perkins ND, Gilmore TD:Good cop, badcop: the different faces of NF-
 B. Cell Death and Differentiation 2006, 13: 759-772. 10.1038/sj.cdd.4401838
B. Cell Death and Differentiation 2006, 13: 759-772. 10.1038/sj.cdd.4401838Rayet B, Gélinas C: Aberrant rel/nfkb genes and activity in human cancer. Oncogene 1999, 18(49):6938-6947. 10.1038/sj.onc.1203221
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Sarkar, A., Meila, M. & Franza, R.B. I B, NF-
B, NF- B Regulation Model: Simulation Analysis of Small Number of Molecules.
J Bioinform Sys Biology 2007, 25250 (2008). https://doi.org/10.1155/2007/25250
B Regulation Model: Simulation Analysis of Small Number of Molecules.
J Bioinform Sys Biology 2007, 25250 (2008). https://doi.org/10.1155/2007/25250
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1155/2007/25250



 B and rel proteins: evolutionary conserved mediators of immune responses. Annual Review of Immunology 1998, 16: 225-260. 10.1146/annurev.immunol.16.1.225
B and rel proteins: evolutionary conserved mediators of immune responses. Annual Review of Immunology 1998, 16: 225-260. 10.1146/annurev.immunol.16.1.225 B signaling pathways. Science 1994, 264(5156):255-258. 10.1126/science.8146656
B signaling pathways. Science 1994, 264(5156):255-258. 10.1126/science.8146656 B
B functions through direct contacts with the nuclear localization signals and the DNA binding sequences of NF-
functions through direct contacts with the nuclear localization signals and the DNA binding sequences of NF- B. Journal of Biological Chemistry 1998, 273(39):25427-25435. 10.1074/jbc.273.39.25427
B. Journal of Biological Chemistry 1998, 273(39):25427-25435. 10.1074/jbc.273.39.25427 B-NF-
B-NF- B signaling module: temporal control and selective gene activation. Science 2002, 298(5596):1241-1245. 10.1126/science.1071914
B signaling module: temporal control and selective gene activation. Science 2002, 298(5596):1241-1245. 10.1126/science.1071914 B signaling control the dynamics of gene expresion. Science 2004, 306(5696):704-708. 10.1126/science.1099962
B signaling control the dynamics of gene expresion. Science 2004, 306(5696):704-708. 10.1126/science.1099962 B signaling control the dynamics of the gene expression. Science 2005, 308(5718):52. 10.1126/science.1107904
B signaling control the dynamics of the gene expression. Science 2005, 308(5718):52. 10.1126/science.1107904 B signaling control the dynamics of the gene expression. Science 2005, 308(5718):52. 10.1126/science.1107904
B signaling control the dynamics of the gene expression. Science 2005, 308(5718):52. 10.1126/science.1107904 B. Cell Death and Differentiation 2006, 13: 759-772. 10.1038/sj.cdd.4401838
B. Cell Death and Differentiation 2006, 13: 759-772. 10.1038/sj.cdd.4401838