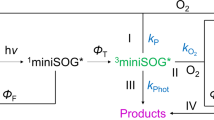
Abstract
miniSOG, developed as the first fully genetically encoded singlet oxygen photosensitiser, has found various applications in cell imaging and functional studies. Yet, miniSOG has suboptimal properties, including a low yield of singlet oxygen generation, which can nevertheless be improved tenfold upon blue light irradiation. In a previous study, we showed that this improvement was due to the photolysis of the miniSOG chromophore, flavin mononucleotide (FMN), into lumichrome, with concomitant removal of the phosphoribityl tail, thereby improving oxygen access to the alloxazine ring. We thus reasoned that a chromophore with a shorter tail would readily improve the photosensitizing properties of miniSOG. In this work, we show that the replacement of FMN by riboflavin (RF), which lacks the bulky phosphate group, significantly improves the singlet oxygen quantum yield (ΦΔ). We then proceeded to mutagenize the residues stabilizing the phosphate group of FMN to alter the chromophore specificity. We identified miniSOG-R57Q as a flavoprotein that selectively binds RF in cellulo, with a modestly improved ΦΔ. Our results show that it is possible to modify the flavin specificity of a given flavoprotein, thus providing a new option to tune its photophysical properties, including those leading to photosensitization. We also determined the structure of miniSOG-Q103L, a mutant with a much increased ΦΔ, which allowed us to postulate the existence of another access channel to FMN for molecular oxygen.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
miniSOG (for mini Singlet Oxygen Generator) is a small flavoprotein derived from the LOV2 (Light, Oxygen and Voltage) domain of Arabidopsis thaliana phototropin 2 that binds flavin mononucleotide (FMN) as a chromophore [1]. It was originally engineered for correlative light and electron microscopy (CLEM) aiming at preserving the fluorescent and photosensitizing properties of FMN. In the last years, miniSOG has been actively used in many applications, often combining advanced microscopy techniques and phototherapy [2].
The ability of miniSOG to generate singlet oxygen (1O2) is, however, low and far from the real capacity of the free FMN molecule in solution (ΦΔ = 0.03 [3, 4] and ΦΔ = 0.51 [5], respectively). Several spectroscopic studies [3, 4, 6,7,8,9] have characterized the optical and photosensitizing properties of miniSOG to examine the factors limiting 1O2 production. Recently, we have reported the crystal structure of miniSOG at high resolution and unveiled the protein and chromophore phototransformations that increase miniSOG’s ΦΔ tenfold, providing final evidence to complete and explain its complex photophysics [10]. The modest ΦΔ is due to an unfavourable combination of efficient FMN deactivation by protein quenching (kp) and the low capacity of molecular oxygen (3O2) to diffuse through the matrix and react with the chromophore (kO2) [7, 10, 11]. Crystallographic data provided the exact distances between the chromophore and the surrounding electron-rich and hydrogen bonding amino acids capable of quenching its triplet excited state (Fig. 1). Moreover, it clearly showed that the isoalloxazine ring of FMN is enclosed in the active pocket and the phosphoribityl tail lies in a tunnel connecting the chromophore to the external media, potentially limiting the oxygen access [10].
Chromophore environment in the 1.17 Å structure of miniSOG. All residues within 4 Å of the FMN are represented. The 2Fobs-Fcalc electron density is represented around the chromophore, the chloride ion and the magnesium-water cluster, contoured at 1.0 σ (blue), 3.0 σ (green) and 5.0 σ (red) levels, highlighting the location of the heavy atoms Cl, Mg, P and the most stable O and N atoms
The design of improved miniSOG variants has mainly focused on suppressing kp. Replacement of key amino acids located in the vicinity of the chromophore effectively boosted ΦΔ values, culminating with the development of SOPP3 (for Singlet Oxygen Photosensitizing Protein version 3, ΦΔ = 0.61) [11]. On the other hand, the contribution of kO2 to the ΦΔ of miniSOG has remained less explored. In the current study, we have sought to improve oxygen accessibility to the chromophore and characterized its effects on 1O2 production and kinetics. We have initially exchanged the FMN molecule by its parental and non-phosphorylated riboflavin (RF) to remove the bulky phosphate group and facilitate oxygen diffusion. In addition, we have rationally developed novel miniSOG mutants binding preferentially RF over FMN and characterized their ability to produce 1O2.
2 Results and discussion
2.1 Replacement of FMN with RF in miniSOG
In our previous work [10], we had shown that the removal of the phosphoribityl tail, which is concomitant to the phototransformation of FMN to lumichrome, as well as the oxidation of electron-rich side chains, contributed to a tenfold increase in ΦΔ. We decided to verify the former assertion by introducing a flavin with a shorter tail than FMN. Vitamin B2, or riboflavin (RF), is a precursor in the biosynthesis of FMN. Lumichrome, however, is not a precursor in RF biosynthesis [12], but a biodegradation product [13], hence its bioavailability is limited, not to mention its poor solubility in water. We thus chose RF as a potential chromophore replacement in miniSOG.
Chromophore replacement was carried out using a denaturation/renaturation protocol (see “Methods”). Briefly, the protein was denatured overnight by incubation with guanidine, leading to the release of FMN, then fixed on a nickel column thanks to its poly histidine tag, and renatured in presence of RF. We used electrospray ionization mass spectrometry (ESI–MS) to assess the successful chromophore replacement (Table 1) and as a quality control before photophysical characterization. We performed time-resolved near-infrared 1O2 phosphorescence spectroscopy to estimate the yield of 1O2 generation on miniSOG, miniSOG reconstituted with RF (miniSOG[RF]), and miniSOG reconstituted with FMN (miniSOG[FMN]) as a control of proper refolding of the protein (Table 2). As hoped, the yield of 1O2 generation was increased 2.5-fold in miniSOG[RF] compared to miniSOG and miniSOG[FMN], supporting our hypothesis that the phosphate group is contributing to restricting 3O2 access to the isolloxazine ring. This is confirmed by the fact that the lifetime of produced 1O2 is approximately constant at ~ 50 μs and that the difference resides in the sixfold reduction of the lifetime of the triplet state of the chromophore (Table 1), suggesting that 3O2 can more readily quench it. Noteworthy, we did not observe any chromophore phototransformation under our irradiation conditions. Because the triplet state plays an important role in the formation of lumichrome in both RF and FMN [14, 15], a more efficient 3O2 quenching of the triplets in miniSOG[RF] reasonably reduces the phototransformation efficiency. We then determined the 1.07 Å structure of miniSOG[RF] and compared it to our previously determined 1.17 Å structure of miniSOG (PDB entry code: 6GPU) [10], to compare the binding modes of FMN and RF. In the structure of miniSOG (Fig. 2a), the phosphate group of FMN is clamped between the side chains of two arginine residues, Arg41 and Arg57, which are conserved in LOV domain-based photosensitizers [9], presumably stabilizing the negative charge(s) of the group. In the structure of miniSOG[RF], however, (Fig. 2b), Arg57 has a completely different conformation, away from FMN, while Arg41 moves sideways, closer to the chromophore (by 0.6 Å for atom Nε, and 0.9 Å for atom Nη2 of the guanidinium group), establishing a hydrogen bond with the terminal O5’ oxygen of the ribityl chain. The movement of Arg41 is correlated with that of Asp14, with which it forms a salt bridge. While this movement slightly narrows the tunnel in miniSOG[RF], the absence of the phosphate group evidently increases the accessibility to the isoalloxazine ring from the bulk solvent (Fig. 2c and d). Interestingly, two chloride ions can be modeled along the ribityl chain of RF in miniSOG[RF] compared to only one next to FMN in miniSOG. As the chloride ion has been shown to act as a mimic of molecular oxygen thanks to the polarizability of its electron cloud [16, 17], these two chloride ions indicate that these locations could serve as transient binding sites for 3O2, between which it could hop before reaching the vicinity of the isoalloxazine ring. All these observations suggest that a miniSOG variant preferentially binding RF could be a better generator of 3O2.
Comparison of the structure of miniSOG with its natural chromophore FMN (miniSOG) or reconstituted with RF (miniSOG[RF]). a Stabilisation of the phosphoribityl tail of FMN in miniSOG. b Stabilisation of the ribityl tail of RF in miniSOG[RF]. c Surface representation of the chromophore-binding pocket in miniSOG with FMN and solvent molecules. d Surface representation of the chromophore-binding pocket in miniSOG with RF and solvent molecules. Water molecules are represented in red, chloride ions in green
2.2 Mutagenesis of miniSOG to favour RF over FMN binding
We reasoned that in order to favour RF binding over FMN, the mutation(s) should destabilize the phosphate group, whose negative charge(s) is(are) stabilized by interactions with two positively charged arginine residues 41 and 57 (Fig. 2a). We mutated both arginines to either glutamine or glutamate, i.e. a residue with a long side chain, which is either neutral or negatively charged. Expression of mutants at position 41 did not result in coloured protein in BL21 cells, contrarily to that of mutants at position 57. This observation can be rationalized by the fact that Arg41 is also in close interaction with RF (Fig. 2b), and that its absence destabilizes the binding of both FMN and RF. In addition, it is very possible that the mutation cancels the electrostatic interaction between Asp14 and Arg41 (on two separate secondary structure elements: the beginning of the second β-strand and the middle of the third α-helix, respectively), which may contribute to stabilizing the chromophore binding site by controlling protein dynamics. We thus expressed and purified miniSOG-R57Q and miniSOG-R57E. Initial 1O2 phosphorescence measurement showed that the R57Q mutant was a slightly superior generator of 1O2 than the R57E mutant and we thus focused on miniSOG-R57Q.
2.3 Characterization of miniSOG-R57Q as a better photosensitizer
We investigated by ESI-mass spectrometry the chromophore content of miniSOG-R57Q when expressed in E. coli. Indeed, the R57Q mutation resulted in an apparent higher affinity of RF binding compared to FMN in a 3:1 ratio (Table 1). We also prepared miniSOG-R57Q denatured and fully renatured with RF for photophysical characterization, a sample dubbed miniSOG-R57Q[RF]. Both proteins show slightly higher values of ΦΔ than miniSOG, consistent with the higher accessibility of oxygen to the chromophore as revealed by the sixfold shorter triplet lifetimes in air-saturated solutions, yet lower than miniSOG[RF] (Table 2). In addition, miniSOG-R57Q[RF] is a slightly better 1O2 generator than miniSOG-R57Q, in line with a slightly shorter lifetime of its triplet state in air-saturated solutions (Table 2), which is consistent with the increased population of RF as the protein chromophore.
We then determined the structures of both miniSOG-R57Q and miniSOG-R57Q[RF]. The latter structure was not obtained from a denatured/renatured protein sample, but by supplementing the expression medium with RF to facilitate protein production for crystallogenesis purpose. The 1.17 Å structure of miniSOG-R57Q[RF] (Fig. 3a) shows a ~ 100% population of RF as a chromophore. In this structure, the stabilization of the RF is very similar to that seen in miniSOG[RF], with a chloride ion and water molecules at the same position around the O5’ terminal atom of the ribityl chain and a hydrogen bond to the Nε atom of Arg41. In both structures, the side chains of residue 57 are orthogonal to the surface of the protein and do not interact with the chromophore. In the structure of miniSOG-R57Q however, the chromophore population is mixed, estimated to 70% FMN vs. 30% RF, in apparent contradiction with the 25% FMN vs. 75% RF population observed in solution as determined by ESI-mass spectrometry. The discrepancy can be explained by a population selection process occurring during crystallogenesis, which is a common purification or enrichment process in chemistry [18]. In this structure, the phosphate group is stabilized by a hydrogen bond from the Nδ2 atom of Gln57 (Fig. 3b), which explains the residual affinity for FMN. The phosphate group is accommodated by a sideways translation of the side chain of Arg41 by a few tens of Angstrom, and the chloride ion seen in both miniSOG[RF] and miniSOG-R57Q[RF] structures is dislodged, probably because of the negative charge(s) of the phosphate group.
Structure of miniSOG-R57Q. a Comparison of the stabilisation of the ribityl tail in miniSOG-R57Q[RF] (magenta) compared to miniSOG[RF] (transparent cyan). b. Stabilisation of the phosphoribityl tail of FMN as observed in the structure of miniSOG-R57Q (dark red) expressed in E. coli in standard conditions
In brief, the R57Q mutation does favour binding of RF over FMN and thus slightly increases oxygen access to the chromophore, hence ΦΔ compared to miniSOG. However, affinity towards FMN is not eliminated, which detracts from realising the full effect of replacing FMN by RF when the protein is expressed in E. coli. Additional mutations are required to completely prevent FMN binding.
2.4 Structure of miniSOG-Q103L
The single-point mutation Q103L on miniSOG, located at the bottom of the FMN-binding pocket, has been shown to be greatly beneficial regarding the yield of 1O2 generation [7]. The variant, dubbed SOPP, shows a sixfold increase in ΦΔ (Table 2), which has been attributed to a decrease in the probability of both photoactivated electron transfer and non-radiative decay of the triplet state via electronic to vibrational energy dissipation [7], as seen from the threefold increase in the triplet lifetime in air-saturated solutions (Table 2). We thus hypothesized that adding the Q103L mutation to our R57Q mutant could result in a significantly improved Type II photosensitizer, as both mutations were responsible for improvement on distinct pathways (increase in oxygen accessibility and reduction of electron transfer). However, in contrast to the single mutant miniSOG-Q103L, we observed by UV–vis absorption spectroscopy that the double mutant miniSOG-R57Q/Q103L was gradually losing its chromophore. We thus determined the structure of miniSOG-Q103L to understand the rationale of this phenomenon.
The 1.37 Å structure of miniSOG-Q103L is very similar to that of miniSOG except on one edge of the isoalloaxazine ring (Fig. 4). Unexpectedly, a water molecule not observed in the structure of miniSOG is apparent on one side of the FMN, bridged through two weak hydrogen bonds to carbonyl groups of Gly40 and FMN (Fig. 4a). This molecule is best modelled with a 50% occupancy, suggesting its mobility. The Q103L mutation has the effect of removing a chain of two successive hydrogen bonds linking the FMN to the surface residue Ser4 via Gln103 (Fig. 4b). These two hydrophilic interactions are replaced in miniSOG-Q103L with a weaker, hydrophobic one, a van der Waals interaction between the phenyl ring of Phe24 and the two methyl groups of Ile103 (Fig. 4b). A consequence of the mutation is the ~ 120° rotation of the Ser4 side chain, which engages in a hydrogen bond with a water molecule of the first solvation sphere of the protein.
Structure of miniSOG-Q103L (SOPP). a 2Fobs-Fcalc electron density contoured at a 0.8 σ level superimposed on the protein model (in yellow) in the vicinity of the chromophore, next to Ile103. A mobile water is visible, weakly hydrogen-bonded to the carbonyl group of Gly40 and to a carbonyl group of the FMN. b. Superposition of the miniSOG (green) and miniSOG-Q103L (yellow) structures illustrating the loss of two hydrogen bonds linking the chromophore to the bulk solvent in miniSOG, and compensated by a van der Waals interaction between Phe24 and Ile103 (in salmon) in miniSOG-Q103L
The implications of the observed structural differences between miniSOG and in miniSOG-Q103L are two-fold. First, the isoalloxazine ring is less stabilized in miniSOG-Q103L than in miniSOG since a strong hydrogen bond to Gln103 is replaced by a weak hydrogen bond to a mobile water molecule, effectively decreasing its stabilisation at this location of the chromophore-binding pocket. On the other hand, the R57Q mutation decreases the affinity for FMN with a weaker stabilisation of the phosphate group or binds RF with less interactions than for FMN. Thus, both mutations contribute to a lesser stabilisation of either FMN or RF, leading to chromophore leakage with time. However, this issue observed during characterization of purified protein samples should not be as problematic in vivo, as the flavin population in cellulo could help replenish at least partially the chromophore content of the double mutant. An in vivo study, such as in Mogensen et al. [19], would help assess the potential of the R57Q/Q103L mutant.
Second, the presence of the water molecule at the bottom of the FMN-binding pocket in miniSOG-Q103L poses the question of solvent accessibility, which is linked to that of oxygen accessibility. The very fact that photosensitization occurs indicate that the isoalloxazine ring is accessible to solvent, which we presumed was through the ribityl tail tunnel [10]. The structure of miniSOG-Q103L suggests that this part of the protein, close to the protein surface, could be more flexible, as the mere disruption of the weak Phe24-Leu103 interaction during protein dynamics may open a solvent channel bridging the bulk solvent and the interior of the protein, allowing access of molecular oxygen and/or exit of 1O2. If indeed this happens in miniSOG-Q103L, it could well happen in miniSOG as well, with the transient disruption of a couple of hydrogen bonds. Finally, the presence of this water molecule in a mixed hydrophobic/hydrophilic environment could also indicate that an affinity site for molecular oxygen has been induced next to the isoalloxazine ring upon the Q103L mutation, a hypothesis that could be tested by pressurizing crystals of miniSOG-Q103L with O2 [20].
3 Conclusion
We have shown that miniSOG reconstituted with RF is thrice as efficient in generating 1O2 compared to miniSOG with is natural chromophore FMN, highlighting RF-binding proteins as attractive potential photosensitisers. We have identified the R57Q mutation as a means to switch the affinity of miniSOG towards RF rather FMN, although incompletely. As a consequence, miniSOG-R57Q is only a marginally better photosensitizer than miniSOG, requiring further improvement. The search for additional mutations led us to characterize the structure of miniSOG-Q103L and to hypothesize that at least one additional entry or exit channel for triplet and singlet oxygen could exist at the bottom of the chromophore binding pocket, in addition to the ribityl tail binding channel.
4 Methods
4.1 Protein mutagenesis, expression and purification
Genes coding for a C-terminal 6xHis-tagged recombinant miniSOG and all miniSOG variants presented in this paper were inserted in a pBAD expression vector (using BamHI/PmeI cloning sites) and over-expressed in Escherichia coli BL21 (DE3) (Invitrogen). The coding sequences of single- and double-point miniSOG mutants were directly synthesized in the pBAD vector containing the miniSOG coding sequence (Genecust, Evry, France). Bacterial cells were grown in 1 L of ZYP-5052 medium containing 100 μg·mL−1 ampicillin at 37 °C, inoculated with 10 mL of an overnight grown starting culture. At an OD600 of 1.0 (± 0.2), expression of recombinant miniSOG was induced by the addition of 0.02% l-arabinose and cells were grown 20 h at 17 °C. Cells were pelleted by centrifugation (4000g, 4 °C, 20 min), re-suspended in 25 mL lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10 mM imidazole, 0.25 mg·mL−1 lysozyme, 400 μg·mL−1 DNAse I, 20 mM MgSO4, 1 tablet of the EDTA-free protease inhibitor cocktail cOmplete (Roche, Basel, Switzerland)) per litre of centrifuged culture. Resuspended pellets were flash-cooled at -80 °C. Thawed pellets were then sonicated (on ice, 6 × 30 s at 35 W power with 30 s resting between each cycle) and cell debris was centrifuged at 15,000g, for 45 min at 4 °C. The protein was purified from the clarified lysate using a nickel affinity column (His-Trap HP 5 ml, GE HealthCare) pre-equilibrated in loading buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10 mM imidazole) and eluted against an imidazole gradient (50 mM Tris pH 8.0, 300 mM NaCl, 10–250 mM imidazole). Fractions of interest were submitted to a last purification step using size-exclusion chromatography (Superdex 75 10/300 GL, GE HealthCare) in 20 mM Tris pH 8.0 buffer, after which the recovered fractions were concentrated to 4 mg·mL−1 (Amicon 4, 10 kDa molecular weight cut-off) for further use. The concentration was determined by UV–vis absorption spectroscopy using a molar absorption coefficient of 14 mM−1·cm−1 at 448 nm. The miniSOG-R57Q[RF] sample used for crystallogenesis was prepared by supplementing the LB medium used to grow E. coli cells with 0.1 g·L−1 of RF (Sigma-Aldrich).
4.2 Chromophore exchange
RF to FMN chromophore exchange was performed on miniSOG and variants using a protocol adapted from a published one [21]. All steps were performed at room temperature. A first step of denaturation of 10 mg of purified miniSOG was performed to remove FMN by overnight incubation under slow stirring on a rotating wheel, in 45 mL of denaturation buffer Pdenat (10 mM Na phosphate pH 8.0, 100 mM NaCl, 6 M Guanidine hydrochloride). The denatured protein is then loaded onto a pre-equilibrated in buffer Pdenat 1 mL HisTrap HP (GE Healthcare) at a low flow rate of 0.2 mL·min−1 to maximize binding efficiency. The column was then washed with 10 column volume (CV) of buffer Pdenat to remove residual FMN. Visual monitoring of yellow color disappearance is used to ensure denaturation. A linear gradient of buffer Pdenat + RF (with [RF] at maximum solubility) to buffer P + RF (10 mM Na phosphate pH 8.0, 100 mM NaCl) from 0 to 100% was set at a flow rate of 0.8 mL·min−1 for at least 30 CV to ensure complete refolding of miniSOG with incorporated RF. The column was then washed with 10 CV of buffer P to remove the RF in excess. A 15 CV gradient was then set to exchange buffer P against buffer A (20 mM Tris pH 8.0, 300 mM NaCl). Renatured miniSOG with RF was then eluted with a gradient of buffer B (20 mM Tris pH 8.0, 300 mM NaCl, 250 mM imidazole). Fractions of interest were then loaded on a desalting column (Hi Trap Desalting 5 mL, GE Healthcare) against buffer C (20 mM tris pH 8.0) and then concentrated (Amicon 4, 10 kDa molecular weight cut-off, Millipore) up to 4 mg·mL−1 for further use.
4.3 Electrospray ionization mass spectrometry (ESI–MS)
Before ESI–MS analysis, the different miniSOG protein samples were diluted to a final concentration of 10 μM in acetonitrile/water/formic acid (50:50:0.1, v/v/v) and infused directly in the mass spectrometer by a syringe pump at a flow rate of 10 μL/min. A blank run was carried out infusing only protein buffer diluted at the same ratio as the protein sample in the same solvent system. Data acquisition was carried out in the positive ion mode with spectra in the profile mode and mass spectra were recorded in the 130–1200 m/z range. The mass spectrometer was operated with the following experimental settings: ESI source temperature was set at 325 °C; nitrogen was used as drying gas (5 L/min) and as nebulizer gas (30 psi); the capillary needle voltage was set at 3500 V. Fragmentor value was of 250 V and skimmer of 65 V. The instrument was operated in the 2 GHz (extended dynamic range) mode and spectra acquisition rate was of 1 spectrum/s. The MS data were acquired and processed with the MassHunter workstation software (Data acquisition v.B.04.00, Qualitative analysis with Bioconfirm v.B.07.00, Agilent Technologies). The relative amount of the two cofactors FMN and RF were estimated by measuring the peak area under the corresponding peaks in ESI mass spectra, i.e. at m/z 457.10 for FMN and at m/z 377.14 for RF, respectively, and the values expressed as area %.
4.4 Photophysical characterization
Spectroscopic measurements were performed using 1 cm quartz cuvettes (Hellma) under magnetic stirring and at room temperature. Absorption and fluorescence spectra were recorded on a double beam Cary 6000i spectrophotometer (Varian) and a Fluoromax-4 spectrofluorometer (Horiba), respectively. 1O2 time-resolved near-infrared (NIR) phosphorescence signals were measured at 1275 nm using a customized PicoQuant Fluotime 200 fluorescence lifetime system. An AO-Z-473 solid state AOM Q-switched laser (Changchun New Industries Optoelectronics Technology Co.) was used for sample excitation at 473 nm. The average power that reached the sample was modulated by neutral density filters. An uncoated SKG-5 filter (CVI Laser Corporation) was placed at the exit port of the laser to remove any residual NIR component. The luminescence exiting from the sample was filtered by a 1100 nm long-pass filter (Edmund Optics) and a narrow bandpass filter at 1275 nm (bk-1270-70-B, bk Interfernzoptik) to remove any scattered laser radiation and isolate the 1O2 emission. A thermoelectric‐cooled NIR sensitive photomultiplier tube assembly (H9170-45, Hamamatsu Photonics) was used as a detector. Photon counting was achieved with a multichannel scaler (NanoHarp 250, PicoQuant Gmbh). The time-resolved 1O2 signals \({S}_{(\mathrm{t})}\) were analyzed by fitting Eq. 1 [22] to the data using GraphPad Prism 5.
where τT and τ∆ are the lifetimes of the photosensitizer triplet state and of 1O2, respectively, and S(0) is a quantity proportional to Φ∆. Flavoprotein Φ∆ values were determined by comparing the S(0) values of optically matched solutions of the corresponding protein and FMN at 473 nm in deuterated buffer (Φ∆FMN = 0.57) [6] as described by Eq. 2.
4.5 Protein crystallisation
miniSOG[RF], miniSOG-R57Q, miniSOG-R57Q[RF] and miniSOG-Q103L were crystallized at a 4 mg.mL−1 concentration, as previously described [10]. Protein crystals appeared in 1–2 days in a condition consisting of 100 mM Tris–HCl pH 8.0, 20 mM MgCl2, 28% PEG 4000, 15 mM CoCl2 at 20 °C.
4.6 Structure determination
X-ray diffraction data were collected at 100 K from flash-cooled crystals that have been cryoprotected with 20% glycerol (v/v) beforehand on beamlines ID23-1 [23] and ID29 [24] of the ESRF in Grenoble (France). Crystallographic data were indexed, integrated, scaled and merged using the XDS package [25]. Our previously determined 1.17 Å structure of miniSOG (PDB entry code: 6GPU) [10], was used as starting model for all structure refinement performed with Refmac5 [26]. Models were manually improved in Coot [27] by reorientation or modification of amino-acid residues and of the flavin chromophores, and addition of solvent molecules. Data collection and refinement statistics are presented in Table 3. Pymol (http://pymol.org/, The PyMOL Molecular Graphics System, Version 1.8.7 Schrödinger, LLC) was used for structural analysis and figure preparation.
Data availability
The structures of miniSOG[RF], miniSOG-R57Q, miniSOG-R57Q[RF] and miniSOG-Q103L are deposited in the Protein Data Bank (http://www.wwpdb.org/) under accession codes 7QF2, 7QF3, 7QF4 and 7QF5.
References
Shu, X., Lev-Ram, V., Deerinck, T. J., Qi, Y., Ramko, E. B., Davidson, M. W., Jin, Y., Ellisman, M. H., & Tsien, R. Y. (2011). A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biology, 9, e1001041. https://doi.org/10.1371/journal.pbio.1001041
Souslova, E. A., Mironova, K. E., & Deyev, S. M. (2017). Applications of genetically encoded photosensitizer miniSOG: From correlative light electron microscopy to immunophotosensitizing. Journal of Biophotonics, 10, 338–352. https://doi.org/10.1002/jbio.201600120
Ruiz-González, R., Cortajarena, A. L., Mejias, S. H., Agut, M., Nonell, S., & Flors, C. (2013). Singlet oxygen generation by the genetically encoded tag miniSOG. Journal of the American Chemical Society, 135, 9564–9567. https://doi.org/10.1021/ja4020524
Pimenta, F. M., Jensen, R. L., Breitenbach, T., Etzerodt, M., & Ogilby, P. R. (2013). Oxygen-dependent photochemistry and photophysics of “MiniSOG,” a protein-encased flavin. Photochemistry and Photobiology, 89, 1116–1126. https://doi.org/10.1111/php.12111
Baier, J., Maisch, T., Maier, M., Engel, E., Landthaler, M., & Bäumler, W. (2006). Singlet oxygen generation by UVA light exposure of endogenous photosensitizers. Biophysical Journal, 91, 1452–1459. https://doi.org/10.1529/biophysj.106.082388
Rodríguez-Pulido, A., Cortajarena, A. L., Torra, J., Ruiz-González, R., Nonell, S., & Flors, C. (2016). Assessing the potential of photosensitizing flavoproteins as tags for correlative microscopy. Chemical Communications, 52, 8405–8408. https://doi.org/10.1039/C6CC03119F
Westberg, M., Holmegaard, L., Pimenta, F. M., Etzerodt, M., & Ogilby, P. R. (2015). Rational design of an efficient, genetically encodable, protein-encased singlet oxygen photosensitizer. Journal of the American Chemical Society, 137, 1632–1642. https://doi.org/10.1021/ja511940j
Westberg, M., Bregnhøj, M., Etzerodt, M., & Ogilby, P. R. (2017). Temperature sensitive singlet oxygen photosensitization by LOV-derived fluorescent flavoproteins. The Journal of Physical Chemistry B, 121, 2561–2574. https://doi.org/10.1021/acs.jpcb.7b00561
Endres, S., Wingen, M., Torra, J., Ruiz-González, R., Polen, T., Bosio, G., Bitzenhofer, N. L., Hilgers, F., Gensch, T., Nonell, S., Jaeger, K. E., & Drepper, T. (2018). An optogenetic toolbox of LOV-based photosensitizers for light-driven killing of bacteria. Scientific Reports, 8, 15021. https://doi.org/10.1038/s41598-018-33291-4
Torra, J., Lafaye, C., Signor, L., Aumonier, S., Flors, C., Shu, X., Nonell, S., Gotthard, G., & Royant, A. (2019). Tailing miniSOG: Structural bases of the complex photophysics of a flavin-binding singlet oxygen photosensitizing protein. Scientific Reports, 9, 2428. https://doi.org/10.1038/s41598-019-38955-3
Westberg, M., Bregnhøj, M., Etzerodt, M., & Ogilby, P. R. (2017). No photon wasted: An efficient and selective singlet oxygen photosensitizing protein. The Journal of Physical Chemistry B, 121, 9366–9371. https://doi.org/10.1021/acs.jpcb.7b07831
Bacher, A., Eberhardt, S., Fischer, M., Kis, K., & Richter, G. (2000). Biosynthesis of Vitamin B2 (Riboflavin). Annual Review of Nutrition, 20, 153–167. https://doi.org/10.1146/annurev.nutr.20.1.153
Lilletvedt, M., Kristensen, S., & Tønnesen, H. H. (2010). Lumichrome complexation by cyclodextrins: Influence of pharmaceutical excipients. Die Pharmazie, 65, 871–876. https://doi.org/10.1691/ph.2010.0680
Schuman Jorns, M., Schöllnhammer, G., & Hemmerich, P. (1975). Intramolecular addition of the riboflavin side chain: Anion-catalyzed neutral photochemistry. European Journal of Biochemistry, 57, 35–48. https://doi.org/10.1111/j.1432-1033.1975.tb02274.x
Sheraz, M. A., Kazi, S. H., Ahmed, S., Anwar, Z., & Ahmad, I. (2014). Photo, thermal and chemical degradation of riboflavin. Beilstein Journal of Organic Chemistry, 10, 1999–2012. https://doi.org/10.3762/bjoc.10.208
Roeser, D., Schmidt, B., Preusser-Kunze, A., & Rudolph, M. G. (2007). Probing the oxygen-binding site of the human formylglycine-generating enzyme using halide ions. Acta Crystallographica Section D, 63, 621–627. https://doi.org/10.1107/S0907444907009961
Colloc’h, N., Gabison, L., Monard, G., Altarsha, M., Chiadmi, M., Marassio, G., Sopkova-De Oliveira Santos, J., El Hajji, M., Castro, B., Abraini, J. H., & Prangé, T. (2008). Oxygen pressurized X-ray crystallography: Probing the dioxygen binding site in cofactorless urate oxidase and implications for its catalytic mechanism. Biophysical Journal, 95, 2415–2422. https://doi.org/10.1529/biophysj.107.122184
Redinha, J. S., Lopes, A. J., Pais, A. A. C. C., & Almei, S. J. A. (2013). Crystallization: From the conformer to the crystal. Advanced Topics on Crystal Growth. https://doi.org/10.5772/54447
Mogensen, D. J., Westberg, M., Breitenbach, T., Etzerodt, M., & Ogilby, P. R. (2021). Stable transfection of the singlet oxygen photosensitizing protein SOPP3: Examining aspects of intracellular behavior. Photochemistry and Photobiology, 2, 1–14. https://doi.org/10.1111/php.13440
Lafumat, B., Mueller-Dieckmann, C., Leonard, G., Colloc’h, N., Prangé, T., Giraud, T., Dobias, F., Royant, A., Van Der Linden, P., & Carpentier, P. (2016). Gas-sensitive biological crystals processed in pressurized oxygen and krypton atmospheres: Deciphering gas channels in proteins using a novel “soak-and-freeze” methodology. Journal of Applied Crystallography, 49, 1478–1487. https://doi.org/10.1107/S1600576716010992
Mansurova, M., Scheercousse, P., Simon, J., Kluth, M., & Gärtner, W. (2011). Chromophore exchange in the blue light-sensitive photoreceptor YtvA from Bacillus subtilis. ChemBioChem, 12, 641–646. https://doi.org/10.1002/cbic.201000515
Nonell, S., & Braslavsky, S. (2000). Time-resolved singlet oxygen detection. Methods in Enzymology (pp. 37–49). Academic Press.
Nurizzo, D., Mairs, T., Guijarro, M., Rey, V., Meyer, J., Fajardo, P., Chavanne, J., Biasci, J. C., McSweeney, S., & Mitchell, E. (2006). The ID23-1 structural biology beamline at the ESRF. Journal of Synchrotron Radiation, 13, 227–238. https://doi.org/10.1107/S0909049506004341
de Sanctis, D., Beteva, A., Caserotto, H., Dobias, F., Gabadinho, J., Giraud, T., Gobbo, A., Guijarro, M., Lentini, M., Lavault, B., Mairs, T., McSweeney, S., Petitdemange, S., Rey-Bakaikoa, V., Surr, J., Theveneau, P., Leonard, G. A., & Mueller-Dieckmann, C. (2012). ID29: A high-intensity highly automated ESRF beamline for macromolecular crystallography experiments exploiting anomalous scattering. Journal of Synchrotron Radiation, 19, 455–461. https://doi.org/10.1107/S0909049512009715
Kabsch, W. (2010). XDS. Acta Crystallographica Section D Biological Crystallography, 66, 125–132. https://doi.org/10.1107/S0907444909047337
Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F., & Vagin, A. A. (2011). REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallographica Section D: Biological Crystallography, 67, 355–367. https://doi.org/10.1107/S0907444911001314
Emsley, P., Lohkamp, B., Scott, W. G., & Cowtan, K. (2010). Features and development of Coot. Acta Crystallographica Section D: Biological Crystallography, 66, 486–501. https://doi.org/10.1107/S0907444910007493
Acknowledgements
The ESRF is acknowledged for access to beamlines and facilities for molecular biology via its in-house research programme. AR acknowledges funding from the French Agence Nationale de la Recherche (project SOxygen, ANR-11-JSV5-0009). SN acknowledges funding from the Spanish Ministerio de Economía y Competitividad (CTQ2016-78454-C2-1-Rand the Fundació la Marató de TV3 (Grant No. 20133133). This work used the mass spectrometry platform of the Grenoble Instruct-ERIC center (ISBG; UMS 3518 CNRS-CEA-UGA-EMBL) within the Grenoble Partnership for Structural Biology (PSB), supported by FRISBI (ANR-10-INBS-0005-02) and GRAL, financed within the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003).
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Pushing the limits of flash photolysis to unravel the secrets of biological electron and proton transfer - a topical issue in honour of Klaus Brettel.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lafaye, C., Aumonier, S., Torra, J. et al. Riboflavin-binding proteins for singlet oxygen production. Photochem Photobiol Sci 21, 1545–1555 (2022). https://doi.org/10.1007/s43630-021-00156-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43630-021-00156-1