Abstract
Background
Human immunodeficiency virus isolates most often use chemokine receptor CCR5 or CXCR4 as a co-receptor to enter target cells. During early stages of HIV-1 infection, CCR5-tropic viruses are the predominant species. The CXCR4-tropic viruses may emerge late in infection. Recognition of factors influencing this phenotypic switch may give some hints on the antiviral strategies like anti-HIV/AIDS drugs, gene therapy and vaccines.
Methods
To investigate the mechanism that triggers R5 to X4 phenotypic switch, we performed a systematic sensitivity analysis based on a five-dimensional model with time-varying parameters. We studied the sensitivity of each factor to the CCR5-to-CXCR4 tropism switch and acquired some interesting outcomes beyond expectation.
Results
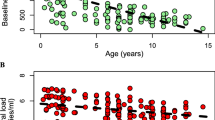
The death rate of free virus (dV), rate that uninfected CD4+ Tcells arise from precursors (s) and proliferate as stimulated by antigens (r), and in vivo viral burst size (N) are four robust factors which are constantly observed to have a strong correlation with the evolution of viral phenotype for most patients longitudinally.
Conclusions
Crucial factors, which are essential to phenotypic switch and disease progression, are almost the same for different patients at different time points, including the production of both virus and CD4+ Tcells and the decay of virion. It is also worth mentioning that although the sequence of factors sorted by the influence varies between patients, the trends of influences engendered by most factors as disease progresses are similar inter-patients.

Article PDF
Similar content being viewed by others
References
Pastore, C., Ramos, A. and Mosier, D. E. (2004) Intrinsic obstacles to human immunodeficiency virus type 1 coreceptor switching. J. Virol., 78, 7565–7574
Koot, M., Keet, I. P. M., Vos, A. H. V., de Goede, R. E. Y., Roos, M. T. L., Coutinho, R. A., Miedema, F., Schellekens, P. T. A. and Tersmette, M. (1993) Prognostic value of HIV-1 syncytiuminducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med., 118, 681–688
Ribeiro, R. M., Hazenberg, M. D., Perelson, A. S. and Davenport, M. P. (2006) Naïve and memory cell turnover as drivers of CCR5-to-CXCR4 tropism switch in human immunodeficiency virus type 1: implications for therapy. J. Virol., 80, 802–809
Swanstrom, R. and Coffin, J. (2012) HIV-1 pathogenesis: the virus. Cold Spring Harb. Perspect. Med., 2, a007443
Sede, M. M., Moretti, F. A., Laufer, N. L., Jones, L. R. and Quarleri, J. F. (2014) HIV-1 tropism dynamics and phylogenetic analysis from longitudinal ultra-deep sequencing data of CCR5- and CXCR4-using variants. PLoS One, 9, e102857
Sguanci, L., Bagnoli, F. and Liò, P. (2007) Modeling HIV quasispecies evolutionary dynamics. BMC Evol. Biol., 7, S5
Moore, J. P., Kitchen, S. G., Pugach, P. and Zack, J. A. (2004) The CCR5 and CXCR4 coreceptors—central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retroviruses, 20, 111–126
van Rij, R. P., Hazenberg, M. D., van Benthem, B. H. B., Otto, S. A., Prins, M., Miedema, F. and Schuitemaker, H. (2003) Early viral load and CD4+ T cell count, but not percentage of CCR5+ or CXCR4+ CD4+ T cells, are associated with R5-to-X4 HIV type 1 virus evolution. AIDS Res. Hum. Retroviruses, 19, 389–398
Boyd, M. T., Simpson, G. R., Cann, A. J., Johnson, M. A. and Weiss, R. A. (1993) A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J. Virol., 67, 3649–3652
Cocchi, F., DeVico, A. L., Garzino-Demo, A., Cara, A., Gallo, R. C. and Lusso, P. (1996) The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat. Med., 2, 1244–1247
Fouchier, R. A. M., Groenink, M., Kootstra, N. A., Tersmette, M., Huisman, H. G., Miedema, F. and Schuitemaker, H. (1992) Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol., 66, 3183–3187
De Jong, J. J., De Ronde, A., Keulen, W., Tersmette, M. and Goudsmit, J. (1992) Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J. Virol., 66, 6777–6780
McKnight, A. and Clapham, P. R. (1995) Immune escape and tropism of HIV. Trends Microbiol., 3, 356–361
Eckstein, D. A., Penn, M. L., Korin, Y. D., Scripture-Adams, D. D., Zack, J. A., Kreisberg, J. F., Roederer, M., Sherman, M. P., Chin, P. S. and Goldsmith, M. A. (2001) HIV-1 actively replicates in naive CD4+ T cells residing within human lymphoid tissues. Immunity, 15, 671–682
Trouplin, V., Salvatori, F., Cappello, F., Obry, V., Brelot, A., Heveker, N., Alizon, M., Scarlatti, G., Clavel, F. and Mammano, F. (2001) Determination of coreceptor usage of human immuno-deficiency virus type 1 from patient plasma samples by using a recombinant phenotypic assay. J. Virol., 75, 251–259
Connor, R. I., Sheridan, K. E., Ceradini, D., Choe, S. and Landau, N. R. (1997) Change in coreceptor use correlates with disease progression in HIV-1–infected individuals. J. Exp. Med., 185, 621–628
van’ t Wout, A. B., Blaak, H., Ran, L. J., Brouwer, M., Kuiken, C. and Schuitemaker, H. (1998) Evolution of syncytium-inducing and non-syncytium-inducing biological virus clones in relation to replication kinetics during the course of human immunodeficiency virus type 1 infection. J. Virol., 72, 5099–5107
Verhofstede, C., Nijhuis, M. and Vandekerckhove, L. (2012) Correlation of coreceptor usage and disease progression. Curr. Opin. HIV AIDS, 7, 432–439
Regoes, R. R. and Bonhoeffer, S. (2005) The HIV coreceptor switch: a population dynamical perspective. Trends Microbiol., 13, 269–277
Tersmette, M. and Miedema, F. (1990) Interactions between HIV and the host immune system in the pathogenesis of AIDS. AIDS, 4, S57–S66
Miedema, F., Tersmette, M. and van Lier, R. A. W. (1990) AIDS pathogenesis: a dynamic interaction between HIV and the immune system. Immunol. Today, 11, 293–297
Berkowitz, R. D., Alexander, S., Bare, C., Linquist-Stepps, V., Bogan, M., Moreno, M. E., Gibson, L., Wieder, E. D., Kosek, J., Stoddart, C. A., et al. (1998) CCR5- and CXCR4-utilizing strains of human immunodeficiency virus type 1 exhibit differential tropism and pathogenesis in vivo. J. Virol., 72, 10108–10117
Le, A. Q., Taylor, J., Dong, W., McCloskey, R., Woods, C., Danroth, R., Hayashi, K., Milloy, M. J., Poon, A. F. Y. and Brumme, Z. L. (2015) Differential evolution of a CXCR4-using HIV-1 strain in CCR5wt/wt and CCR5Δ32/Δ32 hosts revealed by longitudinal deep sequencing and phylogenetic reconstruction. Sci. Rep., 5, 17607
Regoes, R. and Bonhoeffer, S. (2002) HIV coreceptor usage and drug treatment. J. Theor. Biol., 217, 443–457
Perelson, A. S., Kirschner, D. E. and De Boer, R. (1993) Dynamics of HIV infection of CD4+ T cells. Math. Biosci., 114, 81–125
Perelson, A. S. and Nelson, P.W. (1999) Mathematical analysis of HIV-1 dynamics in vivo. SIAM Rev., 41, 3–44
Phillips, A. N. (1996) Reduction of HIV concentration during acute infection: independence from a specific immune response. Science, 271, 497–499
Zhang, Z., Schuler, T., Zupancic, M., Wietgrefe, S., Staskus, K. A., Reimann, K. A., Reinhart, T. A., Rogan, M., Cavert, W., Miller, C. J., et al. (1999) Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science, 286, 1353–1357
Merrill, S. J. (1987) AIDS: Background and the dynamics of the decline of immunocompetence, pp. 59–75. In Theoretical Immunology Workshop
Kupfer, B., Kaiser, R., Rockstroh, J. K., Matz, B. and Schneweis, K. E. (1998) Role of HIV-1 phenotype in viral pathogenesis and its relation to viral load and CD4+ T-cell count. J. Med. Virol., 56, 259–263
Dustin, M. L. and Shaw, A. S. (1999) Costimulation: building an immunological synapse. Science, 283, 649–650
Wilson, D. P., Mattapallil, J. J., Lay, M. D. H., Zhang, L., Roederer, M. and Davenport, M. P. (2007) Estimating the infectivity of CCR5-tropic simian immunodeficiency virus SIV (mac251) in the gut. J. Virol., 81, 8025–8029
Hellerstein, M. K., Hoh, R. A., Hanley, M. B., Cesar, D., Lee, D., Neese, R. A. and McCune, J. M. (2003) Subpopulations of long-lived and short-lived T cells in advanced HIV-1 infection. J. Clin. Invest., 112, 956–966
Hazenberg, M. D., Stuart, J. W., Otto, S. A., Borleffs, J. C. C., Boucher, C. A. B., de Boer, R. J., Miedema, F. and Hamann, D. (2000) T-cell division in human immunodeficiency virus (HIV)-1 infection is mainly due to immune activation: a longitudinal analysis in patients before and during highly active antiretroviral therapy (HAART). Blood, 95, 249–255
McCune, J. M., Hanley, M. B., Cesar, D., Halvorsen, R., Hoh, R., Schmidt, D., Wieder, E., Deeks, S., Siler, S., Neese, R., et al. (2000) Factors influencing T-cell turnover in HIV-1-seropositive patients. J. Clin. Invest., 105, R1–R8
Wu, Y., Lu, Y., Chen, W., Fu, J. and Fan, R. (2012) In silico experimentation of glioma microenvironment development and anti-tumor therapy. PLoS Comput. Biol., 8, e1002355
Acknowledgements
We acknowledge the supports from the National Natural Science Foundation of China (Nos. 11402227, 11621062 and 11432012), the Fundamental Research Funds for the Central Universities of China (No. 2015QNA4034), and the Thousand Young Talents Program of China.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Yu, W., Wu, Y. A systematic analysis of intrinsic regulators for HIV-1 R5 to X4 phenotypic switch. Quant Biol 5, 173–182 (2017). https://doi.org/10.1007/s40484-017-0107-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40484-017-0107-4