Abstract
Key message
Biomass functions are relevant for an easy and quick estimation of tree biomass. Nevertheless, additive biomass functions for different species and different components have not been published for the area of Germany, yet. Now, we present a set of additive biomass functions for estimating component and total mass for eight species and up to nine components.
Context
Biomass functions are relevant for an easy and quick estimation of tree biomass, e.g. for carbon budget calculation. Component-specific functions offer even more detail and can be used to answer questions about, e.g., biomass allocation to different components, (nutrient) element stock and flows or the amount and re-distribution of harvested biomass and its consequences.
Aims
Since there exists no published additive biomass functions in the context of Germany, we aimed at providing such equations for different species and different components using a comprehensive data set from different sources.
Methods
We collected several data sets for eight relevant tree species (Norway spruce, n = 1150 trees; Silver fir, n = 31; Douglas fir, n = 161; Scots pine, n = 460; European beech, n = 918; Oak, n = 313; Sycamore, n = 28 and European ash, n = 37) in Germany and adjacent countries, homogenised the component information, imputed missing values and applied nonlinear seemingly unrelated regression to eight (for deciduous trees species) respectively nine (for conifereous species) components simultaneously.
Results
The collected data set contains trees from 7 cm diameter in breast height to around 80 cm. From this broad data basis, we established two sets of additive biomass functions: a simple model using the predictors diameter in breast height and tree height as well as a more elaborate model using up to six predictors.
Conclusion
Finally, we can present additive models for the eight relevant tree species in Germany. Models for Silver fir, European ash and Sycamore are rather limited in their model range due to their input data; the other models are based on a broad range of predictors and are considered to be broadly applicable.
Similar content being viewed by others
1 Introduction
Information about volume or biomass of trees can easily be derived through allometric equations, as it is done for example for greenhouse gas reporting (IPPC 2003; Oehmichen et al. 2011; de Miguel et al. 2014), for analysis of national forest inventories data (e.g. Riedel and Kaendler 2017) and since many decades in ecological studies (e.g. Marklund 1987). Further applications of biomass functions include research on stem increment, gas-exchange, nutrient and energy flow, forest growth and biomass allocation models (see Wirth et al. 2004; Zianis et al. 2005). Newer applications include the estimation of nutrient export, the amount of available wood for bioenergy and tools for controlling sustainability (Pretzsch et al. 2012; Von Wilpert et al. 2015).
Extensive literature on biomass functions exists (e.g. see Ter-Mikaelian and Korzukhin 1997; Zianis et al 2005), both on total tree or stem biomass (Joosten et al. 2004; Riedel and Kaendler 2017) as well as on component biomass (Pellinen 1986; Marklund 1987; Heinsdorf and Krauß 1990), Cienciala et al.2005, 2006, 2008, Parresol 2001; Wirth et al. 2004; Muukkonen 2007; Wutzler et al. 2008; Krauß and Heinsdorf 2008; Pretzsch et al. 2012; Skovsgaard and Nord-Larsen 2012; de Miguel et al. 2014; Zhao et al. 2015). A wide variety of methods has been applied, both on the natural or logarithmic scale of the data, i.e. nonlinear or linear regression. These include fixed-effects models (e.g. Joosten et al 2004; Riedel and Kaendler 2017), random effects models (e.g. Wirth et al 2004; Wutzler et al 2008) and generalised allometric equations (e.g. Muukkonen 2007; de Miguel et al. 2014). Most studies applied these methods to each biomass component separately, although a desirable trait of component functions is additivity, i.e. that estimates of the component masses sum up to the estimate of the total mass.
Methods which ensure additivity include simple component summation (separately fitted component functions which are additively linked, while variances are summed up and additionally include the covariances between the components, see Parresol 2001), expansion of total or stem estimates using predefined or modelled factors such as specific gravities and proportions (e.g. Forest Inventory and Analysis component ratio method: FIA-CRM, in Poudel and Temesgen 2015; Zell 2008) or biomass expansion factors (BEF, e.g. Lehtonen et al 2004), linear and nonlinear seemingly unrelated regression (Parresol 2001), error-in-variable modelling (Dong et al. 2015) and compositional data analysis (Poudel and Temesgen 2015).
Following Dong et al. (2015), these methods can be divided into aggregative and disaggregative approaches. While most of the procedures work on aggregated data, i.e. the methods start from component functions, where total (or subtotal) mass functions are represented by the sum of the components, compositional data analysis builds on disaggregation, splitting total mass into the respective components via estimated proportions.
In a very recent article of Affleck and Diéguez-Aranda (2016), both approaches have been presented within the maximum-likelihood framework aiming at ‘valid probabilistic models’. By pointing out the additivity property (total mass adds no further information as it is the sum of the components), they argue that in this context the inclusion of total mass leads to invalid probabilistic models and consequently to singularities during estimation. They use the data of and compare results to the aggregative approach of Parresol (2001), showing that they differ only slightly—indeed Parresolachieved slightly smaller standard errors.
To our knowledge, no best method for additive biomass functions has yet emerged, although it could be shown that multivariate methods incorporating intrinsic correlation between components improve on univariate methods (Parresol 2001; Poudel and Temesgen 2015). Comparisons between ‘seemingly unrelated regression’ (SUR) and compositional data analysis (Poudel and Temesgen 2015) or error-in-variable models (Dong et al. 2015) could not show one method being clearly superior to another. Although Poudel and Temesgen (2015) showed that in most of their examples, compositional data analysis led to the lowest RMSE, followed closely by SUR, they did not include the estimation error of the total tree biomass when calculating RMSE for their compositional data models.
The objective of this study is to develop and present a consistent set of additive biomass functions for the most important tree species in Germany, incorporating the correlation between the different components and taking into account the heterogeneity and potential missingness within different components of the different data sources. The ultimate aim of the superior project is to estimate nutrient export during (simulated) harvest to assess sustainability of harvesting operations. Hence, we do not solely focus on statistical aspects, but also provide insights on more practical considerations.
In the following sections, we describe data collection (Section 2.1.1), data harmonisation (Section 2.1.2) and imputation of missing component information (Section 2.1.3). The fitting of component-specific biomass functions is shown in Section 2.2.1, based on which the random effects are estimated and a data adjustment is conducted (Section 2.2.2). Finally, the nonlinear seemingly unrelated regression is described (Section 2.2.3). The results are given in Section 3 also including a comparison to formerly estimated spruce functions (Wirth et al. 2004) and showing the relation between different components. These results are discussed in Section 4, while conclusions are drawn in Section 5.
2 Material and methods
Data preparation and analysis from the raw input data to the final NSUR parameter estimates is described in the following sections and displayed in Fig. 1.
Flow chart for data preparation and analysis. Different data sources (data_1 … n) are consolidated, multiply imputed and fitted by univariate and multivariate procedures. Intermediate steps involve removing grouping structure and deriving regression weights and correlations among components. The final results are formed by pooling the NSUR parameter estimates from each of m = 10 imputed datasets
2.1 Data preparation
2.1.1 Data acquisition
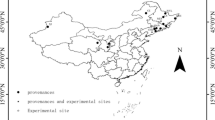
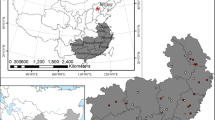
The goal of our study was to develop broadly applicable, additive component biomass functions, and hence, the aim of data acquisition was to gather data from a broad range of environmental conditions. Data were included if the species were of interest and if the geographic extent was suitable (i.e. ‘central Europe’). We could include 17 studies with data mainly from within Germany, but also from neighbouring countries (see Fig. 2 for spruce and Appendix A for all other species). Studies varied in sample size and intention, and also data from other meta-studies were included (e.g. Wirth et al. 2004; Wutzler et al. 2008. Species include Norway spruce (Picea abies (L.) Karst.), Silver fir (Abies alba Mill.), Douglas fir (Pseudotsuga menziesii (Mirb.) Franco) and Scots pine (Pinus sylvestris L.) as well as oak (Quercus robur L. and Q. petraea Liebl.), European beech (Fagus sylvatica L.), European ash (Fraxinus excelsior L.) and Sycamore (Acer pseudoplatanus L.). Although very different component definitions exist, we decided upon a set of additive components commonly recognised in Germany. Firstly, we separated trunk wood and bark. Secondly, coarse wood (i.e. merchantable wood) and bark were split from small wood at 7 cm diameter. For coniferous trees, needles were a separate component. For the final additive biomass models, total aboveground biomass and the summary components ‘trunk including bark’ and ‘coarse wood including bark’ were also included into the models. Abbreviations to these components are given in Table 1. An overview of the collected data is available in Tables 2 and 3.
2.1.2 Data consolidation
The collected data originate from heterogeneous sources with respect to species, sampling methodology, and component selection and definition (e.g. see Wutzler et al. 2008, for different sampling schemes of deciduous tree species). Consolidation of the different studies with respect to component definitions led to a comprehensive list of different components. For example, some studies sampled their trees by separating into main axis and branches, while others sampled according to a threshold diameter of 7 cm, irrespective of branching. While for most studies, the raw data of the sampling were not available or could not be used to calculate further components beside those already given, this was possible with the data from studies 8, 10, 11, 12 and 17 (see Table 2 for references), which made up most of the trees collected. These studies sampled their data either according to the Randomized Branch Sampling (RBS) (Saborowski and Gaffrey 1999; Good et al. 2001) procedure or as full main axis measurement with sampled branches. Consequently, we computed the mass of as many components from the comprehensive component list as possible to remove missingness in the original data. These enhanced raw data, including all available component masses, served as base for the subsequently applied imputation.
2.1.3 Multiple imputation
Missing data is often handled by complete-case analysis (e.g. Wirth et al 2004) or single imputation of conditional or unconditional means; these are methods which potentially exhibit bias or do not take into account the uncertainty of the imputation (Rubin 1987). In contrast, multiple imputation, developed by Rubin (1987), has been shown to treat the deficiencies of these methods while the only ‘extra work’ is to perform the analysis ‘m times instead of once’ (Little and Rubin 2002, p. 86). This is achieved by producing several complete data sets from draws of a predictive distribution, which are subsequently analysed by the desired methods. The results of these methods are finally pooled, e.g. averaged in case of point estimates (see Little and Rubin 2002, for more details).
Missing data methods should be particularly applied when missing data depend on observed variables (confusingly called ‘missing at random’, MAR) or even unobserved, i.e. missing values (‘missing not at random’, MNAR). In contrast, data ‘missing completely at random’ (MCAR) do not lead to biases during estimation. For a more detailed definition, see Little and Rubin (e.g. 2002, p. 11f). At the level of our set of collected data, missingness within each study and components is assumed to be completely at random, but missing components at study level are assumed to be missing by design. This is because at study level it was never intended to record these components. According to Schafer (1997), data missing by design tends to be missing at random and hence should be treated by multiple imputation.
For data preparation and analysis, we used the open source software R (R Core Team 2014). To implement multiple imputation, the mice-package (multivariate imputation by chained equations, van Buuren and Groothuis-Oudshoorn 2011) was used. Imputed values were based on dichotomous component ratios instead of absolute component mass to preserve additivity of all components. Because ratios are bounded within the unit interval, the technique of predictive mean matching (pmm) was chosen, being a ‘general purpose semi-parametric imputation method’ (van Buuren and Groothuis-Oudshoorn 2011, p. 18) which draws new values from the observed values only. Actual component mass was computed by multiplying the respective parent component by the imputed ratio. Additionally, missing predictors were also imputed given the other tree characteristics, but dbh and h were fully observed.
For each tree species, we imputed each missing value ten times (m = 10 chains) and used 30 iterations for the default Gibbs-sampler to obtain the multivariate posterior distribution of our data. Convergence was visually assessed by inspecting the development of the chains, densities of the conditional distributions and the distribution of the imputed values.
The result of the imputation is a set of ten complete data sets. Previously missing observations, which made up 0 to 24 % (up to 47 % in stump components, see further in Table 2 and in Section 3), are now replaced by imputed values, from which the uncertainty of the imputation can be computed at a later stage.
Although the complete set of available components was used during imputation, only the selected set of components—stated in Table 2—were used during model building. Here, multiple imputation not only served the replacement of missing values but also homogenised studies with different sampling scheme.
The effect of the imputation can be measured by the indicator \(\phantom {\dot {i}\!}\lambda \), which gives the proportion of variation attributable to the missing data, i.e. the between-imputation variability divided by the total variability of the parameter estimate. The between-imputation variability is the variance across the m parameter estimates, while the total variability is the sum of the mean of the within-imputation variance (which is usually reported in regression analysis as standard error) and the between-imputation variance. The measure \(\phantom {\dot {i}\!}\lambda \) is given in the results, jointly with the final parameter estimates and corresponding standard errors.
2.2 Data analysis
The method of seemingly unrelated regression (SUR) was invented by Zellner (1962). Parresol explicitly introduced this methodology for nonlinear forest biomass studies using generalised nonlinear least-squares (GNLS) (Parresol 2001). Technically, the multivariate SUR is a system of stacked univariate equations, allowing for different predictors in each equation (block diagonal design matrix) and correlations among the errors of the different responses. To fit nonlinear SUR (NSUR) models, we used code of the systemfit-package (Henningsen and Hamann 2007), extended to allow for regression weights to account for heteroscedasticity. With these extended systemfit-package functions (available on request from the first author), we could reproduce the parameter estimates of the data and models, which are exemplarily given in Parresol (2001) to introduce the NSUR methodology.
2.2.1 Component-specific biomass functions
The NSUR system (Eq. 4 in Section 2.2.3) requires prior knowledge on correlation among components and heteroscedasticity of each component. Hence, we fitted component-specific biomass functions to model heteroscedasticity, to estimate the variance-covariance matrix among the errors of different components, but also to specify model equations and to identify significant predictors. These steps were accomplished using the nlme-package (Pinheiro and Bates 2000).
For each species and component, we fit an allometric model of the form
to find the best-fitting model given a set of predictors. Evaluation criteria were AIC (Akaike 1974) and visual inspection of model diagnostics: Q-Q-plots, residual plots and plots comparing fitted and observed values.
Two sets of predictors were considered: firstly, using dbh and h only (further called ‘simple model’) and, secondly, allowing all significant predictors from the set of dbh, h, a, hsl, upd and cl (henceforth called ‘full model’). Variable cl was considered only for crown components (small wood and needles). For models of stump components, we used sth instead of h as predictor, but included h into the full models additionally if necessary. The product of dbh and h—called dh—was tested if the individual predictors were not sufficient.
Initial tests showed that it is reasonable to model residual variance by a power function to account for the heteroscedastic errors \(\phantom {\dot {i}\!}\epsilon \):
where \(\phantom {\dot {i}\!}\nu \) is a variance covariate and \(\phantom {\dot {i}\!}\delta \) is the variance parameter. In all cases, dbh was used as variance covariate, while some models required a group-specific modelling of the variance to improve on the normality assumption.
Additionally, to account for the factor ‘study’, random effects were tested for all species with a relevant number of groups and trees (excluding Silver Fir, European Ash and Sycamore). The random effects were assumed to be independent of one another. No correlation of errors was assumed at this stage.
2.2.2 Data adjustment
Previous steps of data preparation aimed at unifying components definition and imputing missing values. Still, as the collected data originated from different sources, it is inherently grouped by the factor ‘study’. The method of choice for such data would be to use mixed effects modelling (Pinheiro and Bates 2000, p. 4ff), as we did when modelling the component-specific biomass functions. But as shown in Affleck and Diéguez-Aranda (2016) and further discussed in Section 4, the use of random effects and summary components within a Maximum Likelihood (ML) setting is mutually exclusive.
To account for the grouping, we adjusted the data by removing the estimated ‘study’-effects caused by differences in methodological and technical aspects of data acquisition. From each observation, we subtracted the amount of the prediction, which is attributed to the random effect:
The resulting data, \(\phantom {\dot {i}\!}y_{\text {adj}}\), can be considered to contain only the fixed effects of the raw data. We have checked the difference between the adjusting mixed model on the original data and a structurally identical fixed effect model on the adjusted data by several measures (RMSE = 0.55, MAE = 0.26, BIAS = 0.04, CV = 0.0083, averaged over all models, see Eqs. 5 to 8), and found very small differences only.
2.2.3 Nonlinear seemingly unrelated regression
Seemingly unrelated regression (Zellner 1962) describes a system of regressions, in our case one for each biomass component, whose errors are correlated. Hence, the—in principle independent—regressions become related through their errors’ variance-covariance matrix, making them only ‘seemingly’ unrelated.
Using GNLS, Parresol (2001) showed that in NSUR
minimises the residual sum of squares with respect to the parameter vector \(\phantom {\dot {i}\!}\boldsymbol {\beta }\), where \(\phantom {\dot {i}\!}\boldsymbol {\epsilon }\) are stacked residuals of each equation (𝜖 = Y −f(X,β), with \(\phantom {\dot {i}\!}\boldsymbol {Y}\) being yadj from Eq. 3), \(\phantom {\dot {i}\!}\boldsymbol {{\Delta }}\) is a transformed diagonal weights matrix, Σ is the weighted variance-covariance matrix, incorporating the correlation between different components and \(\phantom {\dot {i}\!}\boldsymbol {I}\) is the unit matrix of dimension n, being the number of observations.
From a theoretical point of view, the advantage of the seemingly unrelated regression is that by using \(\phantom {\dot {i}\!}\boldsymbol {{\Sigma }}\) a system with correlated errors is transformed into one with uncorrelated errors (Zellner 1962; Rossi et al. 2005), allowing the transformed model to be estimated by Least Squares (LS). In case of heteroscedastic errors, the diagonal matrix \(\phantom {\dot {i}\!}\boldsymbol {{\Delta }}\) weights each observation, so that more precise observations get more weight during parameter fitting.
The matrix \(\phantom {\dot {i}\!}\boldsymbol {{\Delta }}\) is the result of an ‘elementwise square root operation on the inverse of the diagonal weights matrix’ (\(\boldsymbol {\Delta } = \sqrt {\boldsymbol {\Psi }^{-1}}\), c.f. Parresol 2001, p. 871), which contains the weights of the fitted component functions. Weights for total and subtotal mass were derived by summing the residuals of the respective components and equivalently modelled by the variance functions of Eq. 2.
The variance-covariance matrix \(\phantom {\dot {i}\!}\boldsymbol {\Sigma }\) was estimated according to Parresol (2001) from the weighted residuals of the univariate component models. Again, the total and subtotal component was obtained by analysing the sum of the residuals of the respective components.
The same model equations as in the component models were used in the multivariate NSUR case. Respective component equations were additively linked to form the model equation for the subtotal (stump wood incl. bark and coarse wood incl. bark) and total aboveground mass. If parameters were insignificant within NSUR and subsequent pooling, they respective predictors were removed from the system of equations, except the scaling parameters and intercepts (if they were necessary for the inclusion of important predictors or proper convergence).
Parameter estimates were obtained by minimising Eq. 4 by means of an optimisation algorithm. Proper convergence was assured by assessing the convergence indicator of the minimisation function (function nlm in stats-package), graphical output of observed and fitted values and of the residuals.
We averaged the results of the ten imputation runs for all of the imputed data sets to receive the final parameter estimates, significance tests and approximate p values according to the pooling rules of Little and Rubin (2002).
2.2.4 Fit statistics
Several fit statistics were calculated for each species and component, subtotal and total mass as the mean of each analysed imputation. These are root mean squared error (RMSE, kg), mean error (BIAS, kg), mean absolute error (MAE, kg) and the coefficient of variation of the RMSE (CV, unitless). These quantities were calculated for each imputed data set and subsequently averaged:
where n is the number of observations, \(\phantom {\dot {i}\!}Y_{i}\) is the i th observation, \(\phantom {\dot {i}\!}\hat {Y}_{i}\) is the i th fitted value and \(\bar {Y}\) is the mean of the observed values.
3 Results
3.1 Additive biomass functions
We included a large number of sample trees for Norway spruce (n = 1150), Douglas fir (n = 161), Scots pine (n = 460), European beech (n = 918) and Oak (n = 313), while the remaining three species had a more limited number of sample trees (Silver fir: n = 31, European ash: n = 37 and Sycamore: n = 28). Our data collection exhibited missingness besides full observations in variable extend, depending on species and component (see Table 2). Highest missingness occured in stump components ranging from 10 to 47 %. Missingness was lowest in aboveground biomass (2 to 15 %). Coarse wood components (10 to 24 %) and crown components (5 to 17 %) showed intermediate values.
The overall minimum dbh of 7 cm reflects the minimum diameter of merchantable wood in Germany. Maximum dbh is at around 80 cm for all species, except for Sycamore with 56 cm and Oak with 95 cm. All but Silver fir, Sycamore and European ash distribute nicely inside a wide range interval for all given variables. For the last three, predictor range and geographical extent are rather limited.
Data adjustment (Section 2.2.2) was conducted for 42 of the 56 univariate mixed effects biomass component models (5 species, 5/6 components, 2 model types). Biggest changes occurred in stump (n = 15), bark (n = 8) and in crown components (n = 9). The adjustment in coarse wood was very small. Notably, Douglas fir exhibited only three adjusted components, all of which were stump components.
The final parameter estimates together with standard error and p-value are given in Table ?? for Norway spruce and in Appendix B for all other species. The corresponding equations are given in Tables 5 and 6 for all species. Additionally, in Table ??, the amount of variability due to missingness is indicated by \(\phantom {\dot {i}\!}\lambda \). For Norway spruce, \(\phantom {\dot {i}\!}\lambda \) is high for stump components, as the rate of missingness is high (c. f. Table 2). For the other components, \(\phantom {\dot {i}\!}\lambda \) often is rather low in the simple models and slightly higher for the full models, especially for crown components. This is because dbh and h are fully observed and with additional incomplete predictors the variability of the parameter estimates increases.
The variability measured by \(\phantom {\dot {i}\!}\lambda \) is not only dependent on the predictor and response variables of the respective component, but also on the other components and their predictors and responses because all components are connected through the variance-covariance-matrix of the errors. An example for this behaviour is the Douglas fir data (Table 10 in Appendix B), where missingness exists only in small wood and needle components, but \(\phantom {\dot {i}\!}\lambda \) is different from zero for all components.
The estimated correlation between different components is high for stem parts and total aboveground biomass as well as for components and their (sub)total aggregates (see Table 7 for Norway spruce, upper triangular matrix). Moderate correlations were found between crown components and between crown and total aboveground biomass. Similar pattern can be seen for all species, with highest correlations between aboveground biomass and total coarse wood. These correlations are accounted for by seemingly unrelated regression. In Appendix C, results for the other tree species are shown.
The full models generally improve over the simple models in terms of RMSE, CV and MAE for all species and components (Tables 5 and 6). BIAS is usually small compared to the RMSE and \(\phantom {\dot {i}\!}\bar {Y}\) ( = RMSE/CV ), with some exceptions in stump and crown components. This is more pronounced in deciduous than in conifer species, especially for stump components of European beech and oak. The relative measure CV indicates, that RMSE is smaller than the mean observation \(\phantom {\dot {i}\!}\bar {Y}\). Only in one case (simple model of needle component for Scots pine) it reaches a value of 1.00. The components cww, cwb and agb exhibit the smallest values (0.1–0.3), while the more variable stump and crown components tend to reach values between 0.3 and 0.7 (in some cases up to 0.9). The pooled variance parameter \(\phantom {\dot {i}\!}\delta \) (see Eq. 2) and the model equations are also included in Tables 5 and 6.
Plotting standardised residuals against the fitted values indicates homoscedasticity and no trend in the residuals (for the full model of Norway spruce see Fig. 3). For bark and stump components (e.g. stw, stb, stwb and cwwb), we encountered some deviations from the normality assumption, finding slightly more positive than negative residuals.
Residual plot of the Norway spruce first imputed data set. The graph plots the the standardised residuals against fitted values for each component. The model fits the data well and the variance model leads to homoscedastic residuals. Slightly more residuals are positive, because negative residuals are bounded to the difference between the mean response and zero—a fact which is also visible, e.g. in Zhao et al. (2015)
The model equations of the simple models rarely changed in the NSUR setting compared to the initial setup from the component-specific functions. Due to their relevance, dbh and h were usually included into the model. In one case—coarse wood bark of ash—the model was constrained on the product of dbh and h ( = dh). In the case of Silver fir, European ash and Sycamore, only dbh was used in crown components. Intercepts were necessary in 11 of 44 cases to improve on normality assumptions. Sometimes, estimated values of intercepts were negative, so care has to be taken in applications.
The full models extended the simple models. For all species but Silver fir, cl was a significant predictor for the crown components. For all conifer species but Silver fir, upd was included for stem and crown components. Upd even replaced dbh for some species and components.
The predictors hsl and a were less important and jointly included only four times, with no instance in deciduous species. Also, both predictors could not be linked to specific components or species, besides that Sycamore included neither, and in contrast, Silver fir and Douglas fir functions each included four instances of hsl and two of a.
In summary, additional predictors were more important for coniferous species, which included 1.2 to 1.6 additional predictors (averaged per species over all components) in comparison to the simple model, while deciduous species added only 0.4 to 1.2 additional predictors.
The pooled residual variance \(\phantom {\dot {i}\!}\sigma _{NSUR}\) (sum of squared errors divided by degrees of freedom) of each system of equations is smaller for the full models compared to the simple models, except for Douglas fir (see Table 8). The highest values of \(\phantom {\dot {i}\!}\sigma _{NSUR}\) are found for Sycamore, European ash and Silver fir, being double to sixfold increased. This indicates that more observations included to the models lead to less residual variance.
In the case of Douglas fir, the small increase in \(\phantom {\dot {i}\!}\sigma _{NSUR}\) can be attributed to the moderate decrease in the sum of the squared errors with respect to ten added parameters to the system of equations.
3.2 Biomass allocation
For all species, biomass allocation changes dramatically between components until 20–30 cm dbh (Fig. 4). Small trees (dbh > 7 cm) exhibit almost equal proportions of coarse wood and small wood. Two exceptions are Sycamore and Silver fir (but both with few empirical data), the first having a coarse wood proportion of about 70 % already for small trees, the latter having only about 30 %. Larger trees, i.e. above 30 cm dbh, are rather constant in their proportions, although species-specific differences exist.
Biomass allocation to components expressed as proportions of total aboveground biomass for each species based on the simple models. Tree height was assumed to follow a Pettersen model (\( h = 1.3 + \left (a + \frac {b}{dbh}\right )^{-3} \)) and fitted by generalised nonlinear regression to the collected observations of each species and stump height was set to be 1 % of tree height. We predicted height for each dbh from 7 to 80 cm, calculated stump height and applied our simple model (the full models would have required more input variables). Results for deciduous trees are given in the top row, for coniferous tree species in the bottom row
Most tree species keep increasing their proportion of coarse wood also beyond 30 cm dbh, mainly drawing from shares of small wood and needles. Sycamore exhibits a rather constant pattern, while European ash keeps changing its proportion with a stronger gradient even for larger trees. In general, coarse wood (including stump mass of about 3–5 %) claims about 80 % of aboveground biomass for trees with dbh\(\phantom {\dot {i}\!}>\) 30 cm. Interestingly, Silver fir reaches the least amount of coarse wood with about 70 %, while Norway spruce is slightly reducing its coarse wood proportion for large trees. In contrast, small wood of Norway spruce tends to increase its proportions for large trees again, while coarse wood bark and needles reduce their relative amount. In other words, the increase in mass given dbh is steeper for small wood than for the other components. This is rather unexpected and one explanation could be that some branches, which as a whole are considered as belonging to small wood, surpassing the 7 cm limit and artificially increasing this component. This might be incorrect with respect to the components size limit, but not for the actual possible stem harvesting mass. The same effect occurs using the models of Wirth et al. (2004). Usually, bark makes up about 10 % of total mass, slightly more in case of Silver fir and Oak, slightly less for Norway spruce and Scots pine. A noticeable exception is European beech, with a very low proportion of bark.
Biomass allocation is relevant when considering energy wood removal during harvest. Nutrient element concentrations differ between components and are regularly higher in crown and bark than in wood (e.g. Weis et al. 2009). As the harvesting of wood is the main interest in forestry, the additional export of nutrients through the inherently connected components should carefully be weighed against the productivity of the respective soils. With these additive biomass functions and in conjunction with nutrient element concentration very detailed harvesting scenarios and optimisations are possible.
3.3 Comparison to other functions
Wirth et al. (2004) developed univariate biomass component functions for Norway spruce in Central Europe. Their raw data were kindly supplied to be included into the presented study (listed as study 9 in Table 1). Figure 5 plots all data from our study, also indicating the data from Wirth et al (2004, black squares) for the three common components coarse wood incl. bark, small wood (branches) and needles. The fitted values of the full model of this study and of the best DHA+C-model (using diameter in breast height, tree height, tree age and crown predictors) of Wirth et al. (2004) are given in the form of a solid and dashed line, respectively, to see differences between both models.
Comparison of our results against the models of Wirth et al. (2004). They fitted their models to trees with no restrictions on dbh, while our data only consists of trees above 7 cm dbh. The solid line represents a smoothing curve based on predictions of the presented NSUR full model, while the dashed smoothing curve is based on the respective best DHA+C-model of Wirth et al (2004, tab. 4, with published correction factors applied). Predictions are based on our data collection
For coarse wood incl. bark, the models seem to be approximately equivalent for \(\phantom {\dot {i}\!}dbh < 70\) cm, while above the line is strongly influenced by only five scattered observations. Additionally, these values lie outside the range of Wirth et al. (2004). The models for small wood and needles exhibit stronger deviations already for small to medium trees and the conditional mean response of the model from Wirth et al. (2004) lies above the conditional mean response of our full models. It seems that the data of Wirth et al. (2004) indicates slightly higher component masses for small wood, especially for medium sized trees, compared to our data. Note that Fig. 5 contains our raw data, with no imputation or adjustment applied, so that data manipulations can be excluded from being the cause of differences.
Predictions for the crown components using the newly fitted functions should be slightly lower and thus more conservative with respect to expected harvest output.
4 Discussion
With this study, we could develop a consistent set of additive biomass functions for eight tree species and eight to nine components, where each component function is based on the full per-species data set. Models for all species were fit using the same procedure, which accounted for intrinsic correlation between components. Raw data were gathered from different sources so that a general applicability of the equations is given for five of eight species at least for the area of Germany. Although fitted the same way, models for Silver fir, European Ash and Sycamore are less general due to limited data availability and geographic extent of the data. Thickest trees show a dbh of more than 80 cm, which is sufficient for most applications. For conservation, where even larger trees are of interest (see e.g. Lutz et al 2012; Stagoll et al 2012, and references therein), this high limit might be important.
Consolidation of the heterogenous data sources was possible because extremely differenciated information was available for some studies and hence component definition of other studies could be mimicked. Remaining missingness in components were handled by multiple imputation. Thereby, we could join studies with different sampling schemes and hence take advantage of the information of as many sample trees as possible.
The model equations are based on allometric theory (Fehrmann and Kleinn 2006), which is well accepted and most commonly applied in literature (c. f. Zianis et al 2005; Ter-Mikaelian and Korzukhin 1997), but alternative parametrisations exist. Main differences between models occur in large trees, where for example Marklund (1987) is more conservative and deviations can be substantial.
Besides hsl, which can be considered tree- or stand-specific, we used only tree-specific predictors to model biomass. \(\phantom {\dot {i}\!}Dbh\) was most frequently used as independent variable, followed by tree height. Indeed, dbh is the easiest and most relevant measure in tree volume and biomass applications, but from a theoretical viewpoint it would be better to use a diameter in relative height (Fehrmann and Kleinn 2006). To support these assertions, we found that although upd and dbh are correlated they were often used jointly, while upd even replaced dbh in some cases. Additional predictors helped to increase accuracy in predictions as could be shown in this study, but also e.g. by Wirth et al. (2004) and Wutzler et al. (2008). Especially crown length (cl) was valuable for the modelling of crown components. Because possible applications might differ in their availability of input variables, we developed two sets of biomass functions (simple and full). The more comprehensive full models, taking advantage of more predictors, usually exhibit less residual variance as well as lower RMSE and CV. Also, BIAS is small with some exceptions in stump and crown components, indicating that the allometric model might not be the ideal functional form to model these components. An additional intrinsic systematic bias like in log-linear models (Sprugel 1983) could be avoided by fitting the models on the data scale.
To fit a system of biomass equations simultaneously, several approaches were described and discussed in literature (e.g. Parresol 2001; Maltamo et al 2012; Dong et al 2015; Poudel and Temesgen 2015; Affleck and Diéguez-Aranda 2016). In this particular case, we had to handle three main difficulties: (i) several studies with different sampling schemes and missing data, (ii) grouping structure of the data, and imperatively and (iii) the inclusion of summary components into the NSUR model. This last requirement excluded models using ML, as decribed by Affleck and Diéguez-Aranda (2016). On the other hand, random effect structures, usually fit by ML are not available in the LS setting (Fahrmeir et al. 2009, p. 263). Hence, we built a data stream from our collected raw data, across enhanced, imputed and adjusted data to pooling of NSUR models fit with GNLS. On that basis, we could include all collected data, incorporate the uncertainty of missing data and treat the grouping of the data. Additionally, through the use of imputation, we avoided potentially biased parameter estimates.
The effect of the data adjustment was a change in variance of the data. We assume that the main effect of the adjustment is in correcting for differences in sampling, since the data adjustment with relevant magnitude mainly occurred in bark and stump components, which are both difficult to sample. Total bark mass can be estimated based on volume or surface area, i.e. interpolated from either measured or modelled bark thickness and assumed shape of the trees. Also, stump components are challenging to measure in the field due to their irregular shape. Hence, larger deviations between different studies are to be expected. Here, the adjustment was used to obtain observations cleared from study specific effects, because the desired response of our models is the conditional mean response of the population average (and not study specific predictions).
By stepping back from the inclusion of summary components, more advanced models such as ML would be possible. ML is able to handle hierarchical data and missingness in the response variable (by allowing unbalanced designs). The question of data harmonisation still applies (c.f. Affleck and Diéguez-Aranda 2016) and fitting parsimonious mixed effect models (Bates et al. 2015) with as many parameters as presented here is not straight forward. In such complex models, the different parts (random effects, correlation and heteroscedasticity) compete with each other (Pinheiro and Bates 2000, p. 204), leading to difficulties in optimisation.
5 Conclusions
Our approach to fit additive biomass functions led to a consistent set of additive nonlinear biomass functions for the most relevant tree species in Germany. We considered a high number of observations, intrinsic correlations between different components and corrected for study effects. Different studies have been included, of which not all component information was available from the raw data. Some studies did not even sample the same components. With our approach, taking advantage of higly differenciated data and multiple imputation, we could consolidate all studies into one scheme of additive components. For historic data, such measures are necessary to be able to use this data but for future data, projects like ENFIN or Cost E43 help harmonising definitions and methods already during acquisation.
From these multiple sets of complete data, we have developed simple and full allometric models applying nonlinear seemingly unrelated regression (NSUR). These models can now be used to compute component mass for trees of interest. A step further, biomass can be transformed into nutrient element mass of different components using respective elemental concentrations. One can also deduce proportions of the different components and their variation with changing predictors.
Although quite some effort has been put into the development, one could still improve estimates by including even larger trees, to better capture the behaviour and variance of the models. Additional observations could improve the models for some species, while for Norway spruce, Scots pine, Douglas fir, European beech and Oak the number of observations seems to be sufficient for model building.
Data availability
The datasets analysed during the current study are available from the corresponding author on reasonable request.
References
Affleck DLR, Diéguez-Aranda U (2016) Additive nonlinear biomass equations: a likelihood-based approach. For Sci 62:129–140. https://doi.org/10.5849/forsci.15-126
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Control 19:716–723. https://doi.org/10.1109/TAC.1974.1100705
Bates D, Kliegl R, Vasishth S, Baayen H (2015) Parsimonious mixed models. ArXiv e-prints p 27. http://adsabs.harvard.edu/abs/2015arXiv150604967B
van Buuren S, Groothuis-Oudshoorn K (2011) Mice: multivariate imputation by chained equations in R. J Stat Softw 45:1–67. https://doi.org/10.18637/jss.v045.i03
Cienciala E, Černý M, Apltauer J, Exnerová Z (2005) Biomass functions applicable to European beech. J For Sci (Prague) 51:147–154
Cienciala E, Černý M, Tatarinov F, Apltauer J, Exnerová Z (2006) Biomass functions applicable to Scots pine. Trees 20:483–495. https://doi.org/10.1007/s00468-006-0064-4, trees
Cienciala E, Apltauer J, Exnerova Z, Tatarinov F (2008) Biomass functions applicable to oak trees grown in Central-European forestry. J For Sci (Prague) 54:109–120
Dong L, Zhang L, Li F (2015) A three-step proportional weighting system of nonlinear biomass equations. For Sci 61:35–45. https://doi.org/10.5849/forsci.13-193
Ellenberg H, Mayer R, Schauermann J (1986) Ökosystemforschung - Ergebnisse des Sollingprojekts : 1966-1986; 145 Tab. Ulmer, Stuttgart, [Hrsg.] Nebent.: Ökosystemforschung - Ergebnisse des Sollingprojekts / Ellenberg ; Mayer ; Schauermann
Fahrmeir L, Lang S, Kneib T (2009) Regression. Springer, Berlin. https://doi.org/10.1007/978-3-642-01837-4
Fehrmann L, Kleinn C (2006) General considerations about the use of allometric equations for biomass estimation on the example of Norway spruce in central Europe. For Ecol Manag 236:412–421. https://doi.org/10.1016/j.foreco.2006.09.026
Good NM, Paterson M, Brack C, Mengersen K (2001) Estimating tree component biomass using variable probability sampling methods. J. Agric. Biol. Environ. Stat. 6:258–267
Heinsdorf D, Krauß HH (1990) Schätztafeln für Trockenmasse und nährstoffspeicherung von kiefernbeständen, IFE-berichte aus Forschung und Entwicklung / Institut für Forstwissenschaften vol 18. Inst. für Forstwiss., Eberswalde-Finow
Henningsen A, Hamann JD (2007) Systemfit: A package for estimating systems of simultaneous equations in R. J. Stat. Softw. 23:40. https://doi.org/10.18637/jss.v023.i04
IPPC (2003) Good practice guidance for land use, Land-Use change and forestry institute for global environmental strategies. IGES, Kanagawa Prefecture
Joosten R, Schumacher J, Wirth C, Schulte A (2004) Evaluating tree carbon predictions for beech (Fagus sylvatica L.) in Western Germany. For Ecol Manag 189:87–96. https://doi.org/10.1016/j.foreco.2003.07.037
Krauß HH, Heinsdorf D (2008) Herleitung von Trockenmassen und nährstoffspeicherungen in buchenbeständen, Eberswalder forstliche Schriftenreihe, vol 38 Ministerium für ländliche Entwicklung. Umwelt und Verbraucherschutz des Landes Brandenburg, Potsdam
Lehtonen A, Makipaa R, Heikkinen J, Sievanen R, Liski J (2004) Biomass expansion factors (BEFs) for scots pine, Norway spruce and birch according to stand age for boreal forests. For Ecol Manag 188:211–224. https://doi.org/10.1016/j.foreco.2003.07.008
Little RJA, Rubin DB (2002) Statistical analysis with missing data, 2nd edn. Wiley series in probability and statistics, Wiley, Hoboken
Lutz JA, Larson AJ, Swanson ME, Freund JA (2012) Ecological importance of large-diameter trees in a temperate mixed-conifer forest. PLoS One 7:e36,131. https://doi.org/10.1371/journal.pone.0036131. http://www.ncbi.nlm.nih.gov/pubmed/22567132
Maltamo M, Mehtätalo L, Vauhkonen J, Packalén P (2012) Predicting and calibrating tree attributes by means of airborne laser scanning and field measurements. Can J For Res 42:1896–1907. https://doi.org/10.1139/x2012-134
Marklund L (1987) Biomass functions for Norway spruce (Picea abies (L.) Karst) in Sweden Report. Department of Forest Survey, SLU
de Miguel S, Mehtätalo L, Durkaya A (2014) Developing generalized, calibratable, mixed-effects meta-models for large-scale biomass prediction. Can J For Res 44:648–656. https://doi.org/10.1139/cjfr-2013-0385
Muukkonen P (2007) Generalized allometric volume and biomass equations for some tree species in Europe. Eur J For Res 126:157–166. https://doi.org/10.1007/s10342-007-0168-4
Oehmichen K, Demant B, Dunger K, Grüneberg E, Hennig P, Kroiher F, Neubauer M, Polley H, Riedel T, Rock J, Schwitzgebel F, Stümer W, Wellbrock N, Ziche D, Bolte A (2011) Inventurstudie 2008 und Treibhausgasinventar Wald. Landbauforschung, Sonderheft, 343
Parresol BR (2001) Additivity of nonlinear biomass equations. Canad J Forest Res-Revue Canadienne De Recherche Forestiere 31:865–878. https://doi.org/10.1139/cjfr-31-5-865
Pellinen P (1986) Biomasseuntersuchungen im Kalkbuchenwald. Thesis
Pinheiro JC, Bates DM (2000) Mixed-effects models in S and S-plus statistics and computing. Springer, New York ; Berlin ; Heidelberg [u.a.] https://doi.org/10.1007/b98882
Poudel KP, Temesgen H (2015) Methods for estimating aboveground biomass and its components for Douglas-fir and lodgepole pine trees. Can J For Res, 77–87. https://doi.org/10.1139/cjfr-2015-0256
Pretzsch H, Göttlein A, Block J (2012) Entscheidungsstutzungssystem zum nährstoffentzug im Rahmen der Holzernte - Teil 1: Textteil.̈ Report Lehrstuhl f. Department Ökosystem- u. Landschaftsmanagement Techn. Univ. München, Waldwachstumskunde
R Core Team (2014) R: a language and environment for statistical computing. http://www.R-project.org/
Riedel T, Kaendler G (2017) Nationale Treibhausgasberichterstattung: Neue Funktionen zur schatzung̈ der oberirdischen Biomasse am Einzelbaum. Forstarchiv 88:31–38
Rossi P, Allenby G, McCulloch R (2005) Bayesian statistics and marketing. Wiley, New York. https://doi.org/10.1002/0470863692
Rubin DB (1987) Multiple imputation for nonresponse in surveys Wiley series in probability and mathematical statistics : Applied probability and statistics. Wiley, New York [u.a.] https://doi.org/10.1002/9780470316696
Rumpf S, Nagel J, Schmidt M (2011) Biomasseschätzfunktionen von Fichte (Picea abies L.), Kiefer (Pinus sylvestris L.) Buche (Fagus sylvatica L.), Eiche (Quercus robur und petraea L.) und Douglasie (Pseudotsuga menziesii L.) für Nordwestdeutschland. Report
Saborowski J, Gaffrey D (1999) RBS, ein mehrstufiges Inventurverfahren zur Schätzung von Baummerkmalen; II. Modifizierte RBS-Verfahren. Allgemeine Forst und Jagdzeitung 170:223– 227
Schafer JL (1997) Analysis of incomplete multivariate data, Monographs on statistics and applied probability, vol 72, 1st edn. Chapman and Hall, London [u.a.]
Schröder J (2014) Biomasseschätzung für Wälder mittels Fernerkundung und Modellierung : Ergebnisse des deutsch-polnischen Verbundprojekts “ForseenPOMERANIA”, Eberswalder forstliche Schriftenreihe, vol 56, 1st edn. Ministerium für Infrastruktur und Landwirtschaft des Landes Brandenburg, [Potsdam], [Red.] Bd. 56 doppelt vergeben
Skovsgaard JP, Nord-Larsen T (2012) Biomass, basic density and biomass expansion factor functions for European beech (Fagus sylvatica L.) in Denmark. Eur J For Res 131:1035–1053. https://doi.org/10.1007/s10342-011-0575-4
Sprugel DG (1983) Correcting for bias in log-transformed allometric equations. Ecol 64:209–210. https://doi.org/10.2307/1937343
Stagoll K, Lindenmayer DB, Knight E, Fischer J, Manning AD (2012) Large trees are keystone structures in urban parks. Conserv Lett 5:115–122. https://doi.org/10.1111/j.1755-263X.2011.00216.x
Ter-Mikaelian MT, Korzukhin MD (1997) Biomass equations for sixty-five North American tree species. For Ecol Manag 97:1–24. https://doi.org/10.1016/S0378-1127(97)00019-4
Von Wilpert K, Vonderach C, Zirlewagen D (2015) Enna - A project for sustainable harvesting wooden biomass. VGB PowerTech 7:83–88
Weis W, Göttlein A (2012) Nährstoffnachhaltige Biomassenutzung. LWF aktuell 90:44–47
Weis W, Gruber A, Huber C, Göttlein A (2009) Element concentrations and storage in the aboveground biomass of limed and unlimed Norway spruce trees at höglwald. Eur J For Res 128(5):437–445. https://doi.org/10.1007/s10342-009-0291-5
Westermann T (2014) Untersuchung auftretender Biomasseverluste entlang der Erntekette bei der Energieholzernte im Buchenholz (Fagus sylvatica L.)
Wirth C, Schumacher J, Schulze ED (2004) Generic biomass functions for Norway spruce in Central Europe - a meta-analysis approach toward prediction and uncertainty estimation. Tree Physiol 24:121–139. https://doi.org/10.1093/treephys/24.2.121
Wutzler T, Wirth C, Schumacher J (2008) Generic biomass functions for Common beech (Fagus sylvatica) in Central Europe: predictions and components of uncertainty. Canad J Forest Res-Revue Canadienne De Recherche Forestiere 38(6):1661–1675. https://doi.org/10.1139/x07-194
Zell J (2008) Methoden für die Ermittlung, Modellierung und Prognose der Kohlenstoffspeicherung in Wäldern auf Grundlage permanenter Großrauminventuren. Thesis
Zellner A (1962) An efficient method of estimating seemingly unrelated regressions and tests for aggregation bias. J Am Stat Assoc 57(298):348–368. https://doi.org/10.2307/2281644
Zhao D, Kane M, Markewitz D, Teskey R, Clutter M (2015) Additive Tree Biomass Equations for Midrotation Loblolly Pine Plantations. Forest Science. https://doi.org/10.5849/forsci.14-193
Zianis D, Muukkonen P, Makipaa R, Mencuccini M (2005) Biomass and stem volume equations for tree species in Europe. Silva Fennica Monograph 1–2:5–63. https://silvafennica.fi/pdf/smf004.pdf
Acknowledgements
We thank Emil Cienciala from Institute of Forest Ecosystem Research, Jílové u Prahy, Czech Republic; Rainer Joosten from the Ministry for Environment, Agriculture, Conservation and Consumer Protection of the State of North Rhine-Westphalia, Düsseldorf, Germany; Simon Klinner from Eberswalde forestry state center of excellence, Eberswalde, Germany; Ralf Moshammer from Chair of Forest Growth and Yield Science, Technical University of Munich, Germany; Sabine Rumpf from Northwest German Forest Research Institute, Göttingen, Germany; Wendelin Weis from Bavarian State Institute of Forestry, Freising, Germany and Christian Wirth from the Department of Systematic Botany and Functional Biodiversity, University of Leipzig, Germany for kindly supplying their data to our analysis.
Funding
This study was funded by BMEL / FNR (FKZ: 22006512).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Aaron R. Weiskittel
Contributions of the co-authors CV collected, processed and analysed the data and wrote the paper.
GK initiated and supported the work by acquiring funding, assisting data analysis and co-editing of the paper.
CFD assisted in data analysis and co-edited the paper.
Appendices
Appendix A: Sample tree origin
Appendix B: Parameter estimates
In the following, the pooled results of the repeated NSUR model fit for the remaining species are presented.
Appendix C: Estimated variance-covariance matrices
The given matrices in this section are pooled from all repeated analyses including both, the within imputation variance and the between imputation variance. On the diagonal (in italics), standard deviations of the errors are given; the lower triangular matrix gives the covariances of the errors between the different components and the upper triangular matrix shows the respective correlations.
Rights and permissions
About this article
Cite this article
Vonderach, C., Kändler, G. & Dormann, C. Consistent set of additive biomass functions for eight tree species in Germany fit by nonlinear seemingly unrelated regression. Annals of Forest Science 75, 49 (2018). https://doi.org/10.1007/s13595-018-0728-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13595-018-0728-4