Abstract
The 2′-substituents distinguish DNA from RNA nucleosides. 2′-O-methylation occurs naturally in RNA and plays important roles in biological processes. Such 2′-modifications may alter the hydrogen-bonding interactions of the nucleoside and thus may affect the conformations of the nucleoside in an RNA chain. Structures of the protonated 2′-O-methylated pyrimidine nucleosides were examined by infrared multiple photon dissociation (IRMPD) action spectroscopy, assisted by electronic structure calculations. The glycosidic bond stabilities of the protonated 2′-O-methylated pyrimidine nucleosides, [Nuom+H]+, were also examined and compared to their DNA and RNA nucleoside analogues via energy-resolved collision-induced dissociation (ER-CID). The preferred sites of protonation of the 2′-O-methylated pyrimidine nucleosides parallel their canonical DNA and RNA nucleoside analogues, [dNuo+H]+ and [Nuo+H]+, yet their nucleobase orientation and sugar puckering differ. The glycosidic bond stabilities of the protonated pyrimidine nucleosides follow the order: [dNuo+H]+ < [Nuo+H]+ < [Nuom+H]+. The slightly altered structures help explain the stabilization induced by 2′-O-methylation of the pyrimidine nucleosides.

Similar content being viewed by others
Introduction
Mass spectrometry has become increasingly popular in the characterization of ribonucleosides [1,2,3]. Qualitative and quantitative analyses via liquid chromatography coupled with mass spectrometry (LC-MS) have contributed significantly to RNA modification analysis [4,5,6,7]. On the generic level, modifications control the activities of the biological system [8]. During standard LC-MS analysis, RNAs are usually digested into nucleosides to facilitate analyses and achieve more consistent results. Mass spectrometry necessarily examines ions rather than neutral molecules, and protonated nucleosides are typically generated and analyzed during LC-MS analysis [4, 5, 7].
Electrospray ionization (ESI) is a soft ionization technique that typically maintains the solution-phase structures of molecules [2]. Thus, the structures of protonated nucleosides in the gas phase are commonly assumed to be the same as found from condensed-phase determinations via NMR and X-ray crystallography. However, these assumptions have generally not been validated by detailed structural investigations in the gas phase. Protonated nucleosides may adopt different sugar conformations in the gas phase due to the lack of solvent and the presence of excess charge. When digested into nucleosides, the phosphate group is cleaved off and the 3′-hydroxy substituent is also free for potential hydrogen-bonding interactions [9]. Further, the presence of the excess proton and the absence of solvent stabilizing hydrogen-bonding interactions may alter the intramolecular hydrogen-bonding interactions and even the tautomeric form of the nucleobase. The gas-phase structures of the protonated and sodium cationized forms of the canonical and some modified DNA and RNA nucleosides have been thoroughly investigated using IRMPD action spectroscopy and electronic structure methods [10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29]. The deprotonated and protonated forms of the canonical DNA and RNA mononucleotides have also been characterized by IRMPD action spectroscopy [30,31,32,33,34,35,36,37,38,39,40] as well as ion mobility mass spectrometry approaches [40, 41], both in conjunction with electronic structure methods. Elucidating the gas-phase structures of the protonated 2′-O-methylated pyrimidine nucleosides, [Nuom+H]+, may facilitate further mass spectral study of modified nucleosides and nucleic acids and provide chemical insight into the structures and folding of oligonucleotides and nucleic acids and their unimolecular decomposition behavior in the gas phase.
Modifications at the 2′ position of the ribose sugar moieties may influence the overall structure and behavior of RNA. Although a large and chemically diverse set of naturally occurring modifications of RNA nucleosides are known, methylation is involved in about two thirds of the known nucleoside modifications [42,43,44]. Each of the canonical RNA nucleobases can undergo modification at several sites; however, modification of the ribosyl moiety has only been found to occur naturally at the 2′ position [45, 46]. 2′-O-methylation is among the naturally occurring modifications, and in particular, 2′-O-methylated pyrimidines are directly involved in gene regulation, especially in the recognition of transfer-RNA (tRNA) [47,48,49,50]. For example, 2′-O-methylcytidine (Cydm) has been found in the anticodon loop of phenylalanine tRNA in yeast, 2′-O-methyuridine (Urdm) is found in glutamine tRNA of Tetrahymena and alanine tRNA of Bombyx mori [51,52,53,54], and 2′-O-methyl-5-methyluridine (Thdm) has been found in lysine and glutamic acid tRNAs from Drosophila and mammals [51, 53, 55].
Methylation of the ribosyl moiety at the 2′ position leads to changes in the electronic and hydrogen-bonding characteristics of the nucleoside [9]. Compared to DNA nucleosides, the presence of the 2′-hydroxy substituent in RNA nucleosides increases the number of favorable rotational orientations of the 2′- and 3′-hydroxy substituents and the potential for canonical and noncanonical hydrogen-bonding interactions [9]. The 2′-O-methyl substituent exhibits hydrogen-bonding characteristics that are intermediate between that of DNA and RNA, as it can act as a hydrogen-bond acceptor, but not a hydrogen-bond donor. As a result, 2′-O-methyl substituents limit the nature of the hydrogen-bonding interactions possible between the 2′- and 3′-hydroxy substituents and noncanonical hydrogen bonding in some low-energy conformers of the RNA nucleosides [10,11,12,13,14]. Thus, the structures of the protonated forms of 2′-O-methylated nucleosides are worthy of study in their own right, but are also compared to the canonical DNA and RNA nucleosides here to elucidate the influence of 2′-O-methylation on their structures and glycosidic bond stabilities.
Structural conformations of 2′-O-methylated pyrimidine nucleosides and nucleotides have been studied using various techniques in the condensed phases [51, 56,57,58,59,60,61,62,63]. The conclusions of these studies consistently indicate that 2′-O-methyl pyrimidine nucleosides adopt 3′-endo sugar puckering and that this modification increases the structural and thermal stability of DNA and RNA strands [51, 56, 61, 64,65,66,67,68,69,70]. However, protonated nucleosides may adopt different sugar conformations in the gas phase due to the lack of solvent and the excess charge.
In the present work, the protonated forms of the 2′-O-methyl pyrimidine nucleosides, [Nuom+H]+, were investigated, including those of 2′-O-methylcytidine, [Cydm+H]+, 2′-O-methyluridine, [Urdm+H]+, and 2′-O-methyl-5-methyluridine, [Thdm+H]+. These nucleoside ions are examined by infrared multiple photon dissociation (IRMPD) action spectroscopy [71,72,73,74] in the IR fingerprint and hydrogen-stretching regions [75,76,77,78,79]. Experiments are supported by theoretical electronic structure calculations to determine the stable low-energy conformers present and their relative stabilities. Further, study of the energy-resolved collision-induced dissociation (ER-CID) behavior of these protonated pyrimidine nucleosides and their DNA and RNA analogues elucidates their relative glycosidic bond stabilities.
Experimental and Computational Methods
Materials
The 2′-O-methylated pyrimidine nucleosides, Cydm and Urdm were purchased from Alfa Aesar (Haverhill, MA, USA), whereas Thdm was purchased from Berry & Associates (Dexter, MI, USA). The canonical DNA and RNA nucleosides, and the 2′-deoxy and 5-methylated nucleosides of uridine were purchased from Sigma-Aldrich (St. Louis, MO, USA). The HPLC grade methanol, water, and formic acid used for IRMPD experiments were purchased from Sigma-Aldrich (Saint-Quentin-Fallavier, France). The HPLC grade methanol used for the ER-CID measurements was purchased from Fischer Scientific (Waltham, MA, USA), and the HPLC grade water was purchased from Sigma-Aldrich (St. Louis, MO, USA). Ammonium acetate used in the ER-CID experiments was purchased from Fluka (St. Louis, MO, USA).
IRMPD Action Spectroscopy Experiments
The 2′-O-methylated pyrimidine nucleosides were dissolved and diluted to 1 mM in a MeOH/H2O (50:50 v/v) mixture with 0.1% formic acid added for the IRMPD measurements in the IR fingerprint region. The IRMPD spectra of their protonated species, [Cydm+H]+, [Urdm+H]+, and [Thdm+H]+, were measured using a 7-T Fourier transform ion cyclotron resonance mass spectrometer (FT-ICR MS, Apex Qe, Bruker Daltonics, Bremen, Germany) coupled to the free electron laser (FEL) at the CLIO user facility [79,80,81]. Ions were generated via electrospray ionization (ESI), mass selected by a quadrupole mass filter, and accumulated in a hexapole ion trap prior to being transmitted to the ICR cell for photodissociation. Detailed instrumental setups have been described previously [79,80,81]. The trapped [Cydm+H]+, [Urdm+H]+, or [Thdm+H]+ ions were irradiated for 220–390 ms with the FEL at 0–5 dB attenuation. Several (typically 3–4) mass scans were summed for IRMPD yield calculations at each IR frequency. For experiments in the hydrogen-stretching region, Cydm, Urdm, and Thdm solutions were diluted to 250 μM in a MeOH/H2O (50:50 v/v) mixture with 0.1% formic acid added. The IRMPD spectra of [Cydm+H]+, [Urdm+H]+, and [Thdm+H]+ were collected on a modified quadrupole ion trap mass spectrometer equipped with an ESI source (LCQ classic, Thermo Finnigan, San Jose, CA, USA) at the University of Lyon [82,83,84]. Ions were irradiated by a YAG-pumped tunable IR optical parametric oscillator/amplifier (OPO) laser system (LaserVision, Bellevue, WA, USA) in the ion trap for 300–400 ms.
IRMPD spectra of [Cydm+H]+, [Urdm+H]+, or [Thdm+H]+ were collected between ~ 900–1900 cm−1 in the fingerprint region and ~ 2850–3700 cm−1 in the hydrogen-stretching region. The IRMPD yield was calculated as the ratio of the sum of the intensities of fragments versus the total ion intensity, Eq. (1),
where \( {I}_{f_i} \) and Ip are the ion intensities of the fragment and precursor ions, respectively. The IRMPD yield was plotted against the laser frequency to generate the final IRMPD spectrum. Intensities were power corrected for the frequency-dependent variations in the laser output. The experimental spectra in the hydrogen-stretching region were plotted as [1-log(IRMPD yield)] to facilitate comparisons between the measured IRMPD and predicted IR spectra.
ER-CID Experiments and Survival Yield Analyses
Energy-resolved collision-induced dissociation (ER-CID) experiments were performed for the protonated forms of the DNA, RNA, and 2′-O-methylated pyrimidine nucleosides on a quadrupole ion trap mass spectrometer (QIT MS, amaZon ETD, Bruker Daltonics, Bremen, Germany) in our laboratory at Wayne State University. The ER-CID method employed has been detailed in previous work reported by our group [17,18,19]. Ions were generated by ESI from solutions containing 10 μM Cydm, Urdm, or Thdm in a MeOH/H2O (50:50 v/v) mixture with 5 mM ammonium acetate. The flow rate of the syringe pump delivering the analyte solution to the ESI source was set at 3 μL/min. The ions were guided into the ion trap, and the [Cydm+H]+, [Urdm+H]+, or [Thdm+H]+ ions were mass isolated, accumulated, and subjected to CID in the ion trap. In addition to providing efficient trapping and cooling of the ions, the helium buffer gas (~ 2 mTorr for enhanced resolution mode) also served as the collision gas for the CID experiments. The qz value of the ion trap for the ER-CID experiments was set to 0.25, which leads to a low-mass cutoff of 27% of the precursor ion m/z. The rf excitation amplitude was increased at a step size of 0.01 V from 0 V to the rf excitation amplitude required to produce complete dissociation of the precursor ions. Experiments were performed in triplicate in order to assess reproducibility. The data were processed using DataAnalysis 4.0 (Bruker Daltonics, Bremen, Germany).
The survival yield was calculated using the intensities of the precursor and fragment ions, as the ratio of the precursor ion intensity to the total ion intensity as described by Eq. (2),
where Ip and \( {I}_{f_i} \) are defined as in Eq. (1). The survival yield was plotted as a function of the rf excitation amplitude. The rf excitation amplitude required to produce 50% dissociation of the precursor ion (CID50%) was extracted by fitting the survival yield curve using the four-parameter logistic dynamic algorithm shown in Eq. (3).
In this equation, max and min are the maximum (1) and minimum (0) values of the survival yield, rfEA is the rf excitation amplitude applied to induce fragmentation, and CIDslope is the slope of the declining region of the survival yield curve. The CID50% values can be correlated with the relative glycosidic bond stabilities when glycosidic bond cleavage is the only CID pathway. CID50% values of the canonical DNA, RNA, and 2′-O-methylated pyrimidine nucleosides were compared to elucidate the effects of 2′-O-methylation on their relative glycosidic bond stabilities. Data analyses were performed using SigmaPlot 10.0 (Systat Software, Inc., San Jose, CA, USA) and custom software developed in our laboratory.
Computational Details
The chemical structures of the 2′-O-methylated pyrimidine nucleosides and several stable low-energy conformers computed for their protonated forms are shown in Figure 1. The most favorable protonation sites of each 2′-O-methylated pyrimidine nucleosides were investigated. N3 and O2 protonation were examined for Cydm, whereas O2 and O4 protonation as well as 2,4-dihydroxy tautomers were considered for Urdm and Thdm based on previous investigations of the canonical DNA and RNA nucleoside analogues [12,13,14]. To generate candidate structures, molecular mechanics simulated annealing procedures [85] were performed using HyperChem software [86] with the AMBER 3 force field. The initial structure for each species was built by modifying (via replacement of the 2′-hydroxy hydrogen atom by a methyl group) the most stable conformers determined for the analogous protonated forms of the canonical RNA nucleosides. Each initial structure was subjected to 0.3 ps of thermal heating, elevating the temperature from 0 to 1000 K. The temperature was then held constant at 1000 K for 0.2 ps to allow sampling of conformational space. The system was then cooled down to 0 K over a period of 0.3 ps. The resulting structure at the end of each cycle was captured and used as the initial structure for the subsequent cycle. This process was repeated for 300 cycles for each protonation site or tautomeric form of Cydm, Urdm, and Thdm examined. Thirty low-energy structures sorted based on the molecular mechanics results were chosen for high-level quantum mechanical calculations. Additional structures were also built and examined to ensure sufficient sampling of the structure pool, and in particular, included the hydroxy and methoxy rotamers of the low-energy structures found after high level calculations. Geometry optimizations and harmonic vibrational frequency analyses were performed using the Gaussian 09 suite of programs [87]. The B3LYP/6-311+G(d,p) level of theory was employed for structural optimization and frequency analysis to provide results directly comparable to those from previous investigations of the analogous protonated DNA and RNA nucleosides. The B3LYP and second-order Møller-Plesset perturbation with full electron correlation, MP2(full), levels of theory with a 6-311+G(2d,2p) basis set were used for single point energy calculations at 298 K, and the relative Gibbs energies of each stable conformer found were extracted and compared [10,11,12,13,14]. The predicted IR spectrum for each optimized structure was also extracted from the calculations. The calculated harmonic frequencies of each conformer were scaled by factors of 0.973–0.980 in the IR fingerprint region, and by 0.957–0.959 in the hydrogen-stretching region before comparison to the experimental IRMPD spectra. In order to reproduce the spectral broadening observed in the experiments, the computed IR spectra were convoluted with a Gaussian line shape having a 20 cm−1 full-width-at-half-maximum (fwhm) in the IR fingerprint region and a 10 cm−1 fwhm in the hydrogen-stretching region.
Chemical structures of the neutral 2′-O-methylated pyrimidine nucleosides are shown. The calculated low-energy conformers N3A and O2A of [Cydm+H]+, O4A, TA, and TB of [Urdm+H]+, and TC, TA, and O2A of [Thdm+H]+, optimized at the B3LYP/6-311+G(d,p) level of theory are shown. The site of protonation or tautomeric state (as indicated by the conformer designation), nucleobase orientation, and sugar puckering are given for each conformer
Results
IRMPD Action Spectroscopy
The major photodissociation pathway observed for all of the protonated 2′-O-methylated pyrimidine nucleosides involves glycosidic bond cleavage with the excess proton retained by the nucleobase, resulting in elimination of the neutral 2′-O-methylated sugar moiety as summarized in reaction (4),
where Nuom is Cydm, Urdm or Thdm, and Nua is the corresponding nucleobase, i.e., cytosine (Cyt), uracil (Ura), or thymine (Thy). The results for [Cydm+H]+ exactly parallel those found for [dCyd+H]+ and [Cyd+H]+ [12] in that reaction (4) is the only dissociation pathway observed. The results for [Urdm+H]+ and [Thdm+H]+ are comparable to those found in the IRMPD experiments of their corresponding protonated DNA and RNA nucleoside analogues, [dUrd+H]+, [Urd+H]+, [dThd+H]+, and [Thd+H]+ [13, 14]. Minor pathways involving water loss are observed in the photodissociation of [dUrd+H]+ and [dThd+H]+. Instead of water loss, the loss of methanol (M) is observed as a very minor fragmentation pathway for both [Urdm+H]+ and [Thdm+H]+ as summarized in reaction (5),
where Nuom is Urdm or Thdm. In addition, glycosidic bond cleavage resulting in loss of the neutral nucleobase from [Urdm+H]+ is also observed as a minor dissociation pathway as described by reaction (6), but was not observed for [dUrd+H]+ or [Urd+H]+.
Due to low signal-to-noise in the experiments for [Urdm+H]+, the very minor dissociation pathway involving loss of methanol was not included in the IRMPD yield calculated in the IR fingerprint region, and only the dominant glycosidic bond cleavage pathway resulting in neutral loss of the sugar was included in the computed IRMPD yield in the hydrogen-stretching region. In both regions, it was confirmed that the IRMPD spectra of these minor products matched that measured for the dominant glycosidic bond cleavage pathway, and that their inclusion in the computed IRMPD yield did not alter the appearance of the spectrum except to make it noisier.
The IRMPD spectrum measured here for [Cydm+H]+ is compared with those measured for the corresponding canonical DNA and RNA analogues, [dCyd+H]+ and [Cyd+H]+, in Figure 2. As can be seen in the figure, the IRMPD spectra of these protonated cytosine nucleoside analogues exhibit highly parallel spectral features. One of the most obvious differences in the IRMPD spectrum of [Cydm+H]+ versus the canonical analogues is that the band at ~ 1790 cm−1 almost disappears. The peak at ~ 1650 cm−1 is also much more intense than other bands nearby; in contrast, the corresponding IR bands of [dCyd+H]+ and [Cyd+H]+ have comparable relative intensities. No IR feature is observed below 1000 cm−1 for [Cydm+H]+, whereas two minor features are observed in the IRMPD spectra of [dCyd+H]+ and [Cyd+H]+ [12]. Splitting of the feature at ~ 3440 cm−1 in the spectrum of [Cydm+H]+ parallels that observed for [Cyd+H]+. The two IR bands near 3550 cm−1 are also fully resolved in [Cydm+H]+, whereas they are not resolved for the canonical DNA and RNA nucleoside analogues.
Comparison of the measured IRMPD spectrum of [Cydm+H]+ to those of its canonical DNA and RNA nucleoside analogues over the IR fingerprint and hydrogen-stretching regions. IRMPD spectra previously reported for [dCyd+H]+ and [Cyd+H]+ are taken from reference [12]
The IRMPD spectrum measured here for [Urdm+H]+ is compared with those measured for the corresponding modified and canonical RNA analogues, [dUrd+H]+ and [Urd+H]+, in Figure 3. Only modest shifts in the band positions are observed in both the IR fingerprint and hydrogen-stretching regions. The band at ~ 1780 cm−1 in the spectrum of [Urdm+H]+ is red shifted by 30 cm−1 compared to those of [dUrd+H]+ and [Urd+H]+. The IR feature at ~ 1100 cm−1 is more intense in the spectrum of [Urdm+H]+ than in the spectra of [dUrd+H]+ or [Urd+H]+. The peak observed at ~ 3400 cm−1 for [Urdm+H]+ is more intense than those observed for [Urd+H]+ and [dUrd+H]+.
Comparison of the measured IRMPD spectrum of [Urdm+H]+ to those of its 2′-deoxy and canonical RNA nucleoside analogues over the IR fingerprint and hydrogen-stretching regions. IRMPD spectra previously reported for [dUrd+H]+ and [Urd+H]+ are taken from reference [13]
The IRMPD spectrum measured here for [Thdm+H]+ is compared with those measured for the corresponding canonical DNA and modified RNA analogues, [dThd+H]+ and [Thd+H]+, in Figure 4. The IRMPD spectra of these protonated thymine nucleoside analogues are also quite parallel. The most obvious spectral difference is that the band observed at ~ 1150 cm−1 for [dThd+H]+ and [Thd+H]+ is not as well resolved, and appears as a shoulder to the blue side of the broad and intense feature at ~ 1100 cm−1, in the spectrum of [Thdm+H]+. Further, the shoulder to the blue of the intense feature at ~ 1200 cm−1 and the moderately intense feature observed at ~ 1380 cm−1 in the spectra of [dThd+H]+ and [Thd+H]+ are much less intense in the spectrum of [Thdm+H]+ such that they are hardly discernible from the baseline. Peak splitting in the spectrum of [Thdm+H]+ at ~ 3560 cm−1 is more obvious than that observed in the spectra of [dThd+H]+ and [Thd+H]+. A very weak shoulder to the blue of these features is observed at ~ 3600 cm−1 for [Thdm+H]+ and [Thd+H]+, but not [dThd+H]+.
Comparison of the measured IRMPD spectrum of [Thdm+H]+ to those of its canonical DNA and 5-methylated RNA nucleoside analogues over the IR fingerprint and hydrogen-stretching regions. IRMPD spectra previously reported for [dThd+H]+ and [Thd+H]+ are taken from reference [14]
ER-CID Experimental Results
As found in the IRMPD experiments, CID of the protonated 2′-O-methylated pyrimidine nucleosides proceeds via a single major fragmentation pathway involving glycosidic bond cleavage with the excess proton retained by the nucleobase, resulting in elimination of the neutral 2′-O-methylated sugar moiety as summarized in reaction (7).
Representative CID mass spectra of the protonated 2′-O-methylated pyrimidine nucleosides measured at rf excitation amplitudes that result in ~ 50% fragmentation are shown in Figure 5. The results for [Cydm+H]+ again parallel those found for [dCyd+H]+ and [Cyd+H]+ (Figure S1), with glycosidic bond cleavage being the only fragmentation pathway observed for these protonated cytosine nucleoside analogues. Methanol (M) loss is also observed as a minor fragmentation pathway for [Urdm+H]+ and [Thdm+H]+, reaction (8). Glycosidic bond cleavage resulting in loss of the neutral nucleobase is observed for [Urdm+H]+ but not for [Thdm+H]+, reaction (9), consistent with the relative proton affinities of the Ura and Thy nucleobases [88,89,90].
Representative CID mass spectra for the protonated uracil and thymine nucleoside analogues are compared in Figures S2 and S3. The CID fragmentation behavior of [Urdm+H]+ and [Thdm+H]+ differ significantly from their canonical and modified DNA analogues. Glycosidic bond cleavage resulting in loss of the neutral nucleobase is not observed for [Urd+H]+, and many water loss pathways are observed for [dUrd+H]+ and [dThd+H]+. Notably, water (W) loss has been replaced by methanol loss in [Urdm+H]+ and [Thdm+H]+, which suggests that the water and methanol losses arise from the ribose moieties. In the case of [dUrd+H]+ and [dThd+H]+, multiple pathways including water loss, even partial ribose loss in [dUrd+H]+ (resulting in the [Ura+W+H]+ fragment ion), and the nucleobase loss all contribute to the survival yield curve. The water loss pathways are dominant over neutral ribose loss in [dUrd+H]+, but not in [dThd+H]+. The higher degree of parallel behavior for the protonated 2′-O-methylated uracil and thymine nucleosides compared to their RNA nucleoside analogues and contrasting with that of their DNA nucleoside analogues suggest that the 2′-hydroxy and 2′-methoxy substituents stabilize the ribose moieties. Further, control experiments performed here suggest that the ketene loss pathway observed previously may have been the result of an impurity in the acetic acid used in those experiments as they were not observed in the current work [17, 18].
Survival Yield Analyses
The survival yield curves measured for the protonated 2′-O-methylated nucleosides are compared to those measured for their canonical DNA and RNA analogues in Figure 6. CID50% values and estimates for the uncertainties in these values extracted from fitting the survival yield data using Eq. (3) are also shown in Figure 6. The reported uncertainties include contributions from the standard deviation in the measurements made in triplicate and the standard error of the mean associated with the least-squares fit. Trends in the CID50% values of the protonated cytosine, uracil, and thymine nucleoside analogues follow the order [dNuo+H]+ < [Nuo+H]+ < [Nuom+H]+, suggesting that 2′-O-methylation strengthens the glycosidic bond relative to the protonated DNA and RNA pyrimidine nucleoside analogues. The observed trend is consistent with absolute activation energies (AEs) measured for glycosidic bond cleavage of the protonated forms of the canonical DNA and RNA pyrimidine nucleosides using threshold CID techniques [89,90,91]. The effect of 2′-O-methylation on the glycosidic bond stability is very slight and of the same order or smaller than the experimental uncertainties in the CID50% values and thus conclusions for [Cydm+H]+ are less robust. Because water losses are the dominant CID pathways for [dUrd+H]+ (see Figure S2), the survival yield curve and CID50% value do not represent the glycosidic bond stability in this case, but instead provide a measure of the overall stability and a lower limit for the glycosidic bond stability. In contrast, the dominant CID pathways of [Urd+H]+ and [Urdm+H]+ do involve glycosidic bond cleavage. Combined with previous accurate thermochemical measurements [89], the glycosidic bond stability order: [dUrd+H]+ < [Urd+H]+ < [Urdm+H]+ is robust. Although water or methanol losses were observed for the protonated thymine nucleoside analogues, ribose loss still dominates the CID behavior of [dThd+H]+, [Thd+H]+, and [Thdm+H]+, see Figure S3. The CID50% value of [Thdm+H]+ slightly exceeds that of [Thd+H]+, and is significantly higher than that of [dThd+H]+, suggesting the glycosidic bond stability follows the order [dThd+H]+ < [Thd+H]+ < [Thdm+H]+.
The conclusions regarding the trends in the glycosidic bond stabilities based on the measured CID50% values are consistent with two-tailed t tests. The p values calculated for each pair of protonated pyrimidine nucleoside analogues are listed in Table S1. The computed p values indicate a > 95% confidence in these trends for all comparisons except [Urd+H]+ vs [Urdm+H]+ (> 91% confidence) and [Cyd+H]+ vs [Cydm+H]+ (only 60% confidence). Clearly accurate thermodynamic measurements of the absolute AEs for glycosidic bond cleavage of the protonated 2′-O-methylated pyrimidine nucleosides are of interest and would help better clarify the energetic differences that this modification produces to the stability of the protonated nucleosides.
Theoretical Results
The stable conformations of [Cydm+H]+, [Urdm+H]+, and [Thdm+H]+ were sampled as described in the “Computational Details” section. Each of the stable B3LYP/6-311+G(d,p) conformers optimized are classified on the basis of the site of protonation or tautomeric form, nucleobase orientation, sugar puckering, and their relative B3LYP/6-311+G(2d,2p) Gibbs energy at 298 K. The B3LYP and MP2(full) methods favor different ground-state structures and protonation sites or tautomeric forms of the protonated canonical pyrimidine nucleoside in several cases [12,13,14]. In the current work, relative Gibbs energies computed at the MP2(full)/6-311+G(2d,2p) level of theory are also provided for more comprehensive evaluation of the energetic trends. The classifications employed for the nucleobase orientation and sugar puckering are described in Figure S4 of the supplementary material [92, 93]. The nucleobase orientation, pseudorotation angle (P), and sugar puckering are calculated based on the methods described previously [15,16,17,18,19, 63]. It is not too surprising that the structures found parallel those of the canonical pyrimidine nucleoside analogues, but exhibit slightly different sugar puckering preferences. The N3A conformer of [Cydm+H]+, Figure 1, is the lowest energy conformer found among all N3 protonated structures computed. The T conformers of [Urdm+H]+ or [Thdm+H]+ represent the 2,4-dihydroxy tautomers, and TA is the lowest energy conformer found among these structures. Tables 1, 2, and 3 list the relative Gibbs energies of select stable low-energy conformers calculated at the B3LYP/6-311+G(2d,2p) and MP2(full)/6-311+G(2d,2p) levels of theory for [Cydm+H]+, [Urdm+H]+, and [Thdm+H]+, respectively. The pseudorotation angle (P), nucleobase orientation, and sugar puckering of each structure are also listed.
Stable Conformations of [Cydm+H]+
Optimized stable B3LYP/6-311+G(d,p) N3 and O2 protonated conformers computed for [Cydm+H]+ are displayed in Figures 1, S5, and S6. The computed results for [Cydm+H]+ are consistent with previous findings for [dCyd+H]+ and [Cyd+H]+, where B3LYP calculations favor N3 protonation over O2, and MP2 calculations suggest a preference for O2 protonation over N3 [12]. However, the energy difference predicted by MP2 for the N3A and O2A conformers is much smaller than that predicted by B3LYP (Table 1). Both N3 and O2 protonated conformers exhibit a preference for an anti nucleobase orientation, whereas conformers with a syn nucleobase orientation are predicted to be less stable. There is an obvious preference for C2′-endo or C3′-endo sugar puckering among all stable structures computed for [Cydm+H]+, with C3′-endo preferred over C2′-endo for the N3 protonated species by ~ 6 kJ/mol, and C2′-endo puckering slightly favored by ~ 1 kJ/mol over C3′-endo for the O2 protonated conformers. Several conformers adopt C3′-exo or C4′-exo sugar puckerings such as conformers N3G and O2I, see Table 1 and Figures S5 and S6. It is not too surprising that these less common sugar puckerings are observed as they are the most similar to C2′-endo and C3′-endo conformations, and characterized by only a modest change in the pseudorotation angle, see Figure S4.
Stable Conformations of [Urdm+H]+
Optimized stable B3LYP/6-311+G(d,p) O2 and O4 protonated conformers as well as 2,4-dihydroxy tautomers (T) computed for [Urdm+H]+ are shown in Figures 1 and S7–S9. The low-energy structures found are quite parallel to those previously found for [dUrd+H]+ and [Urd+H]+ [13]. B3LYP exhibits a preference for O4 protonation, whereas MP2 predicts 2,4-dihydroxy tautomers as being more stable (Table 2). Interestingly, O2 protonated conformers of [Urdm+H]+ are just slightly less energetically favorable, by 8.3 kJ/mol for B3LYP and 5.8 kJ/mol for MP2, compared to the most stable conformers computed. The differences in stability computed for the lowest energy O2 protonated conformers of [dUrd+H]+ and [Urd+H]+ and the corresponding ground conformers are similar or somewhat larger and exceed 7 kJ/mol. The O4 protonated conformers exhibit a preference for an anti nucleobase orientation, whereas the O2 protonated conformers are stabilized by a strong O2H+···O5′ hydrogen-bonding interaction such that the syn orientation is preferred over anti for these conformers. B3LYP predicts that the 2,4-dihydroxy tautomers prefer an anti orientation, whereas MP2 predicts that the 2,4-dihydroxy tautomers prefer a syn orientation. Several O2 protonated conformers and 2,4-dihydroxy tautomers exhibit a strong O2H···O2′ hydrogen-bonding interaction, and these conformers are important in the results for [Urd+H]+. Such O2H···O2′ hydrogen-bonding interactions in [Urdm+H]+ disrupt or at least destabilize the O3′H···O2′ hydrogen-bonding interaction in the sugar moiety. A variety of sugar puckerings are found, with the most common C2′-endo and C3′-endo puckering modes again favored over others. O4 protonated conformers exhibit a preference for C3′-endo over C2′-endo sugar puckering by more than 7 kJ/mol, whereas C2′-endo is favored by less than 2 kJ/mol over C3′-endo for the 2,4-dihydroxy tautomers, and by more than 10 kJ/mol for the O2 protonated conformers.
Stable Conformations of [Thdm+H]+
Optimized stable B3LYP/6-311+G(d,p) O2 and O4 protonated conformers as well as 2,4-dihydroxy tautomers computed for [Thdm+H]+ are shown in Figures 1 and S10–S12. As found for the other protonated 2′-O-methylated nucleosides, the stable conformers calculated for [Thdm+H]+ are highly parallel to those found for [dThd+H]+ and [Thd+H]+ [14]. Although [Thdm+H]+ only differs from [Urdm+H]+ by the 5-methyl substituent, the calculated protonation preference is altered from that of [Urdm+H]+. Both B3LYP and MP2 predict that 2,4-dihydroxy tautomers are the most stable among the structures computed (Table 3). B3LYP suggests that the most stable O2 and O4 protonated conformers are similar in energy and within 2 kJ/mol of the ground 2,4-dihydroxy tautomer. In contrast, MP2 finds larger energy differences among these conformers, especially for the O4A conformer, which is predicted to be 10.0 kJ/mol less stable than the TA conformer. Parallel to the results found for [Urdm+H]+, several O2 protonated conformers and 2,4-dihydroxy tautomers exhibit a strong O2H···O2′ hydrogen-bonding interaction. In contrast to that found for [Thd+H]+, conformers exhibiting a O2H···O2′ hydrogen-bonding interaction are less energetically favored for [Urdm+H]+, especially for the O2 protonated conformers, which is probably related to the disruption of the O3′H···O2′ hydrogen-bonding interaction in the sugar moiety. The O2 protonated conformers and 2,4-dihydroxy tautomers exhibit a preference for a syn nucleobase orientation, whereas the O4 protonated conformers prefer an anti orientation, probably due to the lack of a strong O2H···O5′ hydrogen-bonding interaction. The most common C2′-endo and C3′-endo puckering modes are again favored over all others, with C2′-endo favored over C3′-endo for the O2 protonated conformers and 2,4-dihydroxy tautomers, and C3′-endo preferred over C2′-endo for the O4 protonated species.
Discussion
Conformers of [Cydm+H]+ Populated by ESI
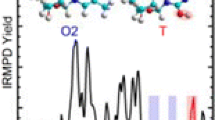
The computed IR spectra for the stable conformers of [Cydm+H]+ are compared with the experimentally measured IRMPD spectrum to elucidate the structures populated in the experiments. The N3A and O2A conformers are found to match the experimental spectrum very well, see the comparison of these spectra shown in Figure 7. The most intense feature at ~ 1660 cm−1 in the IR fingerprint region is associated with C–N stretching and is not diagnostic as it is predicted for both O2 and N3 protonated conformers. N3 protonated conformers provide the signature carbonyl stretch at ~ 1800 cm−1 as well as the major feature at ~ 1660 cm−1 and two small features at ~ 1610 cm−1 and 1560 cm−1, which are related to the C–C or C–N stretches of the cytosine nucleobase. N3A also reproduces the band at ~ 1280 cm−1 very well. In the hydrogen-stretching region, the shoulder to the red of the strong band at ~ 3420 cm−1 arises from the N3–H stretch of N3 protonated conformers. O2A provides the IR feature at ~ 1490 cm−1, which all N3 conformers fail to reproduce. The broad feature observed at ~ 1100 cm−1 is associated with sugar ring breathing, and the shape of the band in the predicted IR spectrum of O2A reproduces this feature better than the N3A conformer. The weak feature predicted for N3A next to the moderately intense feature at 3585 cm−1 represents the O3′–H stretch in the strong O3′H···O2′ hydrogen-bonding interaction. The vibrational frequencies of such hydrogen-bonded modes are often overestimated in harmonic calculations [94,95,96,97]. The strong vibrational band 3585 cm−1 is related to the O2–H stretch of the nucleobase calculated for O2 protonated conformers, which is slightly blue shifted compared to the measured peak. Figures S13 and S14 show similar comparisons for the low-energy N3 and O2 protonated conformers with different sugar puckerings and nucleobase orientations. All of these latter structures exhibit spectral misalignments compared to the experimental spectra such that none of them appear to be significantly populated in the experiments. Detailed vibrational mode assignments based on the N3A and O2A conformers are listed in Table 4. In summary, N3 and O2 protonated conformers of [Cydm+H]+ coexist in the gas phase, with evidence for the N3A and O2A conformers being observed in the experiments.
Comparison of the experimental IRMPD spectrum of [Cydm+H]+ with IR spectra predicted at the B3LYP/6-311+G(d,p) level of theory for the N3A and O2A conformers. The structures, nucleobase orientations, sugar puckering, and B3LYP/6-311+G(2d,2p) (black) and MP2(full)/6-311+G(2d,2p) (red) relative Gibbs energies at 298 K are also listed
Conformers of [Urdm+H]+ Populated by ESI
The experimental spectrum of [Urdm+H]+ is compared with the IR spectra predicted for the O4A, TA, and TB conformers in Figure 8. Figure S15 shows similar comparisons for other conformers (O4F, O4G, O4H, TL, TP, O2N, O2Q, O4P, TY, and TAK) that may be populated in the experiments. However, these latter conformers are less energetically favorable, and do not provide any unique features that can definitively demonstrate their presence in the experiments. The 2,4-dihydroxy tautomers generally reproduce the IR bands at ~ 1640 cm−1, ~ 1500 cm−1, and in the region of ~ 1050–1250 cm−1 very well. In the hydrogen-stretching region, 2,4-dihydroxy tautomers also provide the IR features observed at ~ 3570 cm−1. The TB conformer matches this feature even better than the TA conformer, even though the peak calculated at ~ 1580 cm−1 is slightly red shifted versus the measured feature. Both O4 and O2 protonated conformers may contribute to the carbonyl stretch observed at ~ 1780 cm−1. The IR features in the IR fingerprint region are not sensitive to the difference between these two protonation sites. The band at ~ 3400 cm−1 may arise from either O4 or O2 protonated conformers because that feature represents the N3–H stretch, which is lacking in the tautomers. However, O2A (Figure S18) exhibits misalignment of the IR band observed at ~ 3680 in the hydrogen-stretching region. The peak whose frequency is under-predicted is associated with the O5′–H stretch, which is likely the result of the hydrogen-bonding interaction with the 2-hydroxy substituent. B3LYP and MP2 both predict O2A as being higher in Gibbs energy compared to the O4A and TA conformers. Overall, O4 protonated conformers provide a better match to the experimental data than O2 protonated conformers, suggesting O4 protonation is more important in the experiments. Figures S16–S18 show comparisons of the computed IR and experimental IRMPD spectra for conformers of [Urdm+H]+ that adopt other nucleobase orientations and sugar puckerings. The spectral mismatches are shaded in red. It is worth pointing out that the TC and TE conformers (Figure S17) exhibit O2–H···O2′ hydrogen-bonding interactions. The analogous tautomers are populated significantly in the canonical RNA nucleoside, [Urd+H]+ [13]. However, current results indicate that this conformation exhibits an obvious band shift at ~ 1450 and ~ 1550 cm−1, suggesting that this conformation is not populated for [Urdm+H]+. The experimental IRMPD spectrum clearly arises from contributions from multiple conformers having different protonation sites or tautomeric forms (see Figure 8 and Figure S15). We conclude that the O4A, TA, and TB conformers are significantly populated in the experiments. Among the higher-energy conformers, O4E is the 4-hydroxy rotamer of O4A. O4C also adopts a similar conformation to that of O4A, but the lack of the O3′H···O2′ hydrogen bond makes it less stable than O4A. The nucleobase orientation of O4D is quite parallel to that of O4A; however, both B3LYP and MP2 find C2′-endo sugar puckering much less stable. Detailed vibrational mode assignments based on the O4A, TA, and TB conformers are listed in Table 5.
Comparison of the experimental IRMPD spectrum of [Urdm+H]+ with IR spectra predicted at the B3LYP/6-311+G(d,p) level of theory for the O4A, TA, TB conformers. The structures, nucleobase orientations, sugar puckering, and B3LYP/6-311+G(2d,2p) (black) and MP2(full)/6-311+G(2d,2p) (red) relative Gibbs energies at 298 K are also listed
Conformers of [Thdm+H]+ Populated by ESI
The measured IRMPD and the IR spectra predicted for the TA, O2A, and TC conformers of [Thdm+H]+ are compared in Figure 9. All three conformers have Gibbs energies within 5 kJ/mol of one other and are plausibly populated in the experiments. The TA conformer reproduces the IR feature at ~ 1490 cm−1 very well. The peak shapes between 1050 and 1250 cm−1 also represent the experimental spectrum nicely. The O2A conformer provides the distinguishable carbonyl band at ~ 1780 cm−1 in the fingerprint region and the N3–H stretch at ~ 3410 cm−1 in the hydrogen-stretching region. The TC conformer matches the strong IR band at ~ 3590 cm−1, which is not reproduced by either the TA or O2A conformers. Other conformers that provide a reasonable match to the experimental spectrum are shown in Figure S19. All of these latter conformers are computed to be greater than 20 kJ/mol higher in Gibbs energy than the ground conformer, and none provide any unique IR features that could confirm their presence in the experiments. Figures S20 and S21 show comparisons of the computed IR and experimental IRMPD spectra for O2 protonated conformers and 2,4-dihydroxy tautomers that adopt other nucleobase orientations and sugar puckerings. The spectral mismatches between the predicted and experimental spectra are indicated with red shading. Consistent with [Urdm+H]+, conformers with an O2H···O2′ hydrogen bond (TB, TBG, O2B, O2C, and O2E), cannot be significantly populated in the experiments. All O4 protonated conformers (see Figure S22) exhibit obvious shifts in the carbonyl stretch at ~ 1780 cm−1 and the IR band at ~ 3590 cm−1 that is associated with the O4–H stretch. Unlike [Urdm+H]+, the 5-methyl substituent makes the difference between O2 and O4 protonation more distinguishable in both regions. Overall, O2 protonated conformers and 2,4-dihydroxy tautomers are highly favored structures in the experiments. Conformers TA, O2A, and TC (see Figure 9) contribute to the experimental spectrum significantly, whereas conformers TI, TL, O2N, and TBR (Figure S19) may also contribute. Vibrational mode assignments based primarily on the TA and O2A conformers are listed in Table 6. The unique IR band contributed by the TC conformer is noted.
Comparison of the experimental IRMPD spectrum of [Thdm+H]+ with IR spectra predicted at the B3LYP/6-311+G(d,p) level of theory for the TA, O2A, and TC conformers. The structures, nucleobase orientations, sugar puckering, and B3LYP/6-311+G(2d,2p) (black) and MP2(full)/6-311+G(2d,2p) (red) relative Gibbs energies at 298 K are also listed
Influence of the O2′-Substituent on the Stable Conformations of the Protonated Pyrimidine Nucleosides
The dominant conformers N3A and O2A of [Cydm+H]+, and similar conformers found for [dCyd+H]+ and [Cyd+H]+ are compared in Figure S23 [12, 21]. The nucleobase always adopts an anti orientation, and the sugar puckering is either C2′-endo or C3′-endo. However, N3 and O2 protonated conformers prefer C2′-endo sugar puckering in [dCyd+H]+ and [Cyd+H]+. The N3A conformer of [Cydm+H]+ adopts C2′-endo puckering, whereas the O2A conformer adopts C3′-endo puckering. Theoretically, B3LYP always favors N3 protonation, whereas MP2 prefers O2 protonation. However, in the case of [dCyd+H]+ and [Cyd+H]+, B3LYP and MP2 results only differ by a few kJ/mol [12]. In the case of [Cydm+H]+, the energy difference between N3A and O2A predicted by B3LYP is ~ 7 kJ/mol, whereas MP2 finds these conformers to be within 1 kJ/mol of one another, which seems more realistic on the basis of the experimental IRMPD spectrum.
According to previous work, many conformers of [dUrd+H]+ and [Urd+H]+ are populated by ESI [13]; present results suggest that even more conformers may be present in the experiments for [Urdm+H]+, see Figure 8 and Figure S15. Significantly populated conformers of [Urdm+H]+ are compared to those of [dUrd+H]+ and [Urd+H]+ in Figure S24. The O4 protonated conformers and 2,4-dihydroxy tautomers are dominant for [Urdm+H]+, [dUrd+H]+, and [Urd+H]+. Among all conformers populated significantly, O4 conformers always adopt an anti orientation even though the energy differences between anti and syn oriented tautomers are not large. In [dUrd+H]+ and [Urd+H]+, C2′-endo and C3′-endo sugar puckering do not cause a dramatic change in energy. For [Urdm+H]+, there is a clear preference for sugar puckering based on the site of protonation. The O4 protonated conformers favor C3′-endo sugar puckering (see O4A in Figure S7), and even though there is no hydrogen bond between the 2′- and 3′-hydroxy substituents, the relative energy is still slightly lower than a similar conformer that adopts C2′-endo sugar puckering (compare O4C and O4D, Figure S7). This sugar puckering preference is similar to that found for the N3 protonated conformers of [Cydm+H]+. Interestingly, many tautomers present in the experiments adopt C2′-endo sugar puckering, whereas C3′-endo is not even found among the structures significantly populated. The sugar puckering preference is consistent with O2 protonated conformers of [Cydm+H]+. Lastly, the unique structures that involve O2H···O2′ hydrogen-bonding interactions are only found for [Urd+H]+. Parallel conformers that have hydrogen-bonding interactions between the nucleobase 2-hydroxy and sugar 2′-hydroxy substituents are found theoretically, but those structures are not populated in significant amounts in the experiments. In that regard, [Urdm+H]+ is more similar to [dUrd+H]+.
[Thdm+H]+, [dThd+H]+, and [Thd+H]+ all exhibit a preference for O2 protonated conformers and 2,4-dihydroxy tautomers, and their low-energy conformers are compared in Figure S25 [14, 20]. Structures with O2H···O2′ hydrogen-bonding interactions are only found theoretically for [Thdm+H]+ and are predicted to be energetically competitive with other stable low-energy structures. However, comparison of the measured IRMPD and predicted IR spectra of [Thdm+H]+ suggest that these conformers cannot be populated significantly in the experiments, whereas they are very important for [Thd+H]+. Aside from these O2H···O2′ hydrogen-bonded conformers, the tautomers of [Thdm+H]+, [dThd+H]+, and [Thd+H]+ adopt highly parallel structures. Both syn and anti orientations are found in all three species, and C2′-endo sugar puckering is favored in combination with a syn orientation. The anti oriented tautomers exhibit a preference for C2′-endo sugar puckering for [dThd+H]+ and [Thd+H]+, but prefer C3′-endo sugar puckering for [Thdm+H]+ (see for example the TC conformer). For O2 protonated conformers, it is consistent that a syn nucleobase orientation with C2′-endo sugar puckering is favored for all three thymine nucleoside analogues, with the orientation of the hydrogen-bonding interactions in [Thd+H]+ and [Thdm+H]+ being the only difference.
Comparisons Between [Cydm+H]+, [Urdm+H]+, and [Thdm+H]+
The N3A conformer of [Cydm+H]+, O4A conformer of [Urdm+H]+, and TC conformer of [Thdm+H]+ all adopt anti, C3′-endo conformations. No syn, C3′-endo conformers are populated in any of these species. This is probably related to the sugar rigidity induced by the bulky 2′-O-methyl group. When the sugar adopts C2′-endo puckering, the 2′-O-methyl pyrimidine nucleosides exhibit greater flexibility in the nucleobase orientation. O2A of [Cydm+H]+ and TA of [Urdm+H]+ still adopt an anti orientation with 2T3 sugar puckering. However, TB of [Urdm+H]+ and TA and O2A of [Thdm+H]+ adopt syn nucleobase orientations, and their sugar puckering changes to 2T1. Systematic matching of the nucleobase orientation (anti vs. syn) and preferred C2′-endo puckering (2T3 vs 2T1) is found among all of the pyrimidine nucleosides.
Influence of the O2′-Substituent on the Dissociation Behavior of the Protonated Pyrimidine Nucleosides
As described above, the most favorable dissociation pathway for all of the protonated pyrimidine nucleoside analogues is glycosidic bond cleavage with the excess proton retained by the nucleobase, reaction (4). Based on the mechanisms and absolute energetics for glycosidic bond cleavage established for the canonical DNA and RNA nucleosides [89,90,91], it is clear that the proton affinities (PA) of the pyrimidine nucleobases [88] exceed those of the 2′-deoxyribose and ribose sugar moieties (although PA values have not been reported for the sugars) as glycosidic bond cleavage with the excess proton retained by the sugar, reaction (6) is more energetic by 59.8 kJ/mol for [dCyd+H]+, 63.7 kJ/mol for [Cyd+H]+, 4.8 kJ/mol for [dUrd+H]+, 22.1 kJ/mol for [Urd+H]+, 15.5 kJ/mol for [dThd+H]+, and 18.2 kJ/mol for [Thd+H]+ kJ/mol. Given the very high PA of cytosine 949.9 kJ/mol, it is not surprising that the sugar cannot compete for the excess proton and that no minor neutral loss pathways involving the sugar are observed. As the PAs of uracil and thymine are much lower, 872.7 and 880.9 kJ/mol, respectively, the sugar is able to more effectively compete for the excess proton and the neutral loss pathways become somewhat more important particularly for the dUrd and dThd. Substitution at the 2′-position stabilizes the sugar moiety such that minor neutral loss pathways involving the sugar are almost entirely shut down, but does not influence the PA of the sugar sufficiently to make glycosidic bond cleavage with the excess proton retained by the sugar, reaction (6), very competitive.
Relative Glycosidic Bond Stabilities of Protonated DNA, RNA, and 2′-O-Methylated Nucleosides
The glycosidic bond stabilities follow the order [dNuo+H]+ < [Nuo+H]+ < [Nuom+H]+ for all of the protonated pyrimidine nucleosides, suggesting that 2′-O-methylation slightly strengthens the glycosidic bond relative to the protonated DNA and RNA nucleoside analogues. However, the guanine nucleosides were found to follow the order [dGuo+H]+ < [Guom+H]+ < [Guo+H]+, and the glycosidic bonds of the protonated adenine nucleoside analogues are probably stronger than those of all of the other protonated nucleosides based on the significantly higher CID50% values determined [24]. A detailed understanding of the energetic differences between the protonated 2′-O-methylated purine and pyrimidine nucleosides requires detailed transition state calculations and accurate guided ion beam tandem mass spectrometry measurements, which are currently being pursued by our research group.
Gas-Phase Versus Condensed-Phase Structures
All 2′-O-methylated pyrimidine nucleosides were found to prefer C3′-endo sugar puckering in the condensed phase [51, 63, 98]. The correct codon recognition and thermostability of tRNA was explained as arising from the conformational rigidity of the 2′-O-methylated nucleoside residues, in particular, the inherent stability of the C3′-endo over C2′-endo sugar puckering [51]. In the gas phase, anti, C3′-endo conformers are still present for all of the protonated 2′-O-methylated pyrimidine nucleosides, [Cydm+H]+, [Urdm+H]+, and [Thdm+H]+. It is not surprising that the syn or anti, C2′-endo sugar puckering conformations are competitive with the anti, C3′-endo conformations in the gas phase as a result of the flexibility and the protonation of the 2′-O-methylated pyrimidine nucleosides. On the other hand, the glycosidic bond stability of the 2′-O-methylated pyrimidine nucleosides follow the order [dNuo+H]+ < [Nuo+H]+ < [Nuom+H]+. The results are consistent with the increased thermostability of tRNA and other DNA and RNA studies [51, 56, 66,67,68,69].
Conclusions
IRMPD action spectroscopy experiments and theoretical analyses were performed for the 2′-O-methylated pyrimidine nucleosides to determine the structures accessed in the experiments. The preferred protonation sites or tautomeric forms of the 2′-O-methylated pyrimidine nucleosides parallel their canonical and modified DNA and RNA nucleoside analogues, yet their nucleobase orientations and sugar puckerings differ. The glycosidic bond stabilities of the protonated pyrimidine nucleosides all follow the order [dNuo+H]+ < [Nuo+H]+ < [Nuom+H]+. [Cydm+H]+ adopts an anti, C3′-endo (3T2) configuration in the most stable N3 protonated conformer, N3A, an anti, C2′-endo (2T3) for the most stable O2 protonated conformer, O2A, and these conformers dominate the experimental population. The major contributors to the measured spectrum of [Urdm+H]+ are the O4 protonated, anti, C3′-endo (3T2) conformer, O4A, and the 2,4-dihydroxy tautomers, TA and TB with anti, C2′-endo (2T3) and syn, C2′-endo (2T1) configurations, respectively. For [Thdm+H]+, the O2 protonated, syn, C2′-endo (2T1) conformer, O2A, 2,4-dihydroxy tautomer, syn, C2′-endo (2T1) conformer, TA and anti, C3′-endo (3T2) conformer, TC, contribute to the IRMPD experimental spectrum. The trends in the glycosidic bond stabilities of the protonated DNA, RNA, and 2′-O-methylated pyrimidine nucleosides appear to be related to minor differences in conformation and the nature and variety of hydrogen-bonding interactions available to these species. More accurate thermochemical measurements via threshold CID approaches for the protonated forms of the 2′-O-methylated pyrimidine nucleosides are desirable and are currently being pursued to obtain absolute activation energies for glycosidic bond cleavage, and to definitively establish the influence of 2′-O-methylation on the glycosidic bond stability of the protonated pyrimidine nucleoside analogues.
References
Helm, M., Motorin, Y.: Detecting RNA modifications in the epitranscriptome: Predict and validate. Nat. Rev. Genet. 18, 275–291 (2017)
Banoub, J.H., Limbach, P.A.: Mass spectrometry of nucleosides and nucleic acids. CRC Press, Boca Raton (2010)
Dudley, E., Bond, L.: Mass spectrometry analysis of nucleosides and nucleotides. Mass Spectrom. Rev. 33, 302–331 (2014)
Edmonds, C.G., Crain, P.F., Gupta, R., Hashizume, T., Hocart, C.H., Kowalak, J.A., Pomerantz, S.C., Stetter, K.O., McCloskey, J.A.: Posttranscriptional modification of transfer-RNA in thermophilic archaea (archaebacteria). J. Bacteriol. 173, 3138–3148 (1991)
Noon, K.R., Bruenger, E., McCloskey, J.A.: Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile sulfolobus solfataricus. J. Bacteriol. 180, 2883–2888 (1998)
Guymon, R., Pomerantz, S.C., Crain, P.F., McCloskey, J.A.: Influence of phylogeny on posttranscriptional modification of rRNA in thermophilic prokaryotes: The complete modification map of 16S rRNA of thermus thermophilus. Biochemistry-US. 45, 4888–4899 (2006)
Su, D., Chan, C.T.Y., Gu, C., Lim, K.S., Chionh, Y.H., McBee, M.E., Russell, B.S., Babu, I.R., Begley, T.J., Dedon, P.C.: Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat. Protoc. 9, 828–841 (2014)
Bjork, G.R., Ericson, J.U., Gustafsson, C.E.D., Hagervall, T.G., Jonsson, Y.H., Wikstrom, P.M.: Transfer-RNA modification. Annu. Rev. Biochem. 56, 263–287 (1987)
Nelson, D.L., Cox, M.M.: Lehninger principles of biochemistry, 7th edn. W. H. Freeman and Company, New York (2017)
Wu, R.R., Yang, B., Berden, G., Oomens, J., Rodgers, M.T.: Gas-phase conformations and energetics of protonated 2′-deoxyguanosine and guanosine: IRMPD action spectroscopy and theoretical studies. J. Phys. Chem. B. 118, 14774–14784 (2014)
Wu, R.R., Yang, B., Berden, G., Oomens, J., Rodgers, M.T.: Gas-phase conformations and energetics of protonated 2′-deoxyadenosine and adenosine: IRMPD action spectroscopy and theoretical studies. J. Phys. Chem. B. 119, 2795–2805 (2015)
Wu, R.R., Yang, B., Frieler, C.E., Berden, G., Oomens, J., Rodgers, M.T.: N3 and O2 protonated tautomeric conformations of 2′-deoxycytidine and cytidine coexist in the gas phase. J. Phys. Chem. B. 119, 5773–5784 (2015)
Wu, R.R., Yang, B., Frieler, C.E., Berden, G., Oomens, J., Rodgers, M.T.: Diverse mixtures of 2,4-dihydroxy tautomers and O4 protonated conformers of uridine and 2′-deoxyuridine coexist in the gas phase. Phys. Chem. Chem. Phys. 17, 25978–25988 (2015)
Wu, R.R., Yang, B., Frieler, C.E., Berden, G., Oomens, J., Rodgers, M.T.: 2,4-dihydroxy and O2 protonated tautomers of dThd and Thd coexist in the gas phase: Methylation alters protonation preferences versus dUrd and Urd. J. Am. Soc. Mass Spectrom. 27, 410–421 (2016)
Zhu, Y., Hamlow, L.A., He, C.C., Strobehn, S.F., Lee, J.K., Gao, J., Berden, G., Oomens, J., Rodgers, M.T.: Influence of sodium cationization versus protonation on the gas-phase conformations and glycosidic bond stabilities of 2′-deoxyadenosine and adenosine. J. Phys. Chem. B. 120, 8892–8904 (2016)
Zhu, Y., Hamlow, L.A., He, C.C., Lee, J.K., Gao, J., Berden, G., Oomens, J., Rodgers, M.T.: Gas-phase conformations and N-glycosidic bond stabilities of sodium cationized 2′-deoxyguanosine and guanosine: Sodium cations preferentially bind to the guanine residue. J. Phys. Chem. B. 121, 4048–4060 (2017)
Zhu, Y., Roy, H.A., Cunningham, N.A., Strobehn, S.F., Gao, J., Munshi, M.U., Berden, G., Oomens, J., Rodgers, M.T.: Effects of sodium cationization versus protonation on the conformations and N-glycosidic bond stabilities of sodium cationized Urd and dUrd: solution conformation of [Urd+Na]+ is preserved upon ESI. Phys. Chem. Chem. Phys. 19, 17637–17652 (2017)
Zhu, Y., Roy, H.A., Cunningham, N.A., Strobehn, S.F., Gao, J., Munshi, M.U., Berden, G., Oomens, J., Rodgers, M.T.: IRMPD action spectroscopy, ER-CID experiments, and theoretical studies of sodium cationized thymidine and 5-methyluridine: Kinetic trapping during the ESI desolvation process preserves the solution structure of [Thd+Na]+. J. Am. Soc. Mass Spectrom. 28, 2423–2437 (2017)
Zhu, Y., Hamlow, L.A., He, C.C., Lee, J.K., Gao, J., Berden, G., Oomens, J., Rodgers, M.T.: Conformations and N-glycosidic bond stabilities of sodium cationized 2′-deoxycytidine and cytidine: Solution conformation of [Cyd+Na]+ is preserved upon ESI. Int. J. Mass Spectrom. 429, 18–27 (2018)
Salpin, J.Y., Scuderi, D.: Structure of protonated thymidine characterized by infrared multiple photon dissociation and quantum calculations. Rapid Commun. Mass Spectrom. 29, 1898–1904 (2015)
Filippi, A., Fraschetti, C., Rondino, F., Piccirillo, S., Steinmetz, V., Guidoni, L., Speranza, M.: Protonated pyrimidine nucleosides probed by IRMPD spectroscopy. Int. J. Mass Spectrom. 354, 54–61 (2013)
Ung, H.U., Huynh, K.T., Poutsma, J.C., Oomens, J., Berden, G., Morton, T.H.: Investigation of proton affinities and gas phase vibrational spectra of protonated nucleosides, deoxynucleosides, and their analogs. Int. J. Mass Spectrom. 378, 294–302 (2015)
Abo-Riziq, A., Crews, B.O., Compagnon, I., Oomens, J., Meijer, G., von Helden, G., Kabelac, M., Hobza, P., de Vries, M.S.: The mid-IR spectra of 9-ethyl guanine, guanosine, and 2′-deoxyguanosine. J. Phys. Chem. A. 111, 7529–7536 (2007)
He, C.C., Hamlow, L.A., Devereaux, Z.J., Zhu, Y., Nei, Y.-w., Fan, L., McNary, C.P., Maitre, P., Steinmetz, V., Schindler, B., Compagnon, I., Armentrout, P.B., Rodgers, M.T.: Structural and energetic effects of O2′-ribose methylation of protonated purine nucleosides. J. Phys. Chem. B. 122, 9147–9160 (2018)
Hamlow, L.A., Devereaux, Z.J., Roy, H.A., Cunningham, N.A., Berden, G., Oomens, J., Rodgers, M.T.: Impact of the 2'- and 3′-sugar hydroxyl moieties on gas-phase nucleoside structure. J. Am. Soc. Mass Spectrom. 30, 832–845 (2019)
Hamlow, L.A., He, C.C., Devereaux, Z.J., Roy, H.A., Cunningham, N.A., Soley, E.O., Berden, G., Oomens, J., Rodgers, M.T.: Gas-phase structures of protonated arabino nucleosides. Int. J. Mass Spectrom. 438, 124–134 (2019)
Hamlow, L.A., Zhu, Y., Devereaux, Z.J., Cunningham, N.A., Berden, G., Oomens, J., Rodgers, M.T.: Modified quadrupole ion trap mass spectrometer for infrared ion spectroscopy: Application to protonated thiated uridines. J. Am. Soc. Mass Spectrom. 29, 2125–2137 (2018)
Devereaux, Z.J., Roy, H.A., He, C.C., Zhu, Y., Cunningham, N.A., Hamlow, L.A., Berden, G., Oomens, J., Rodgers, M.T.: Influence of 2′-fluoro modification on glycosidic bond stabilities and gas-phase ion structures of protonated pyrimidine nucleosides. J. Fluor. Chem. 219, 10–22 (2019)
Devereaux, Z.J., Zhu, Y., Rodgers, M.T.: Relative glycosidic bond stabilities of naturally occurring methylguanosines: 7-methylation is intrinsically activating. Eur. J. Mass Spectrom. (Chichester). 25, 16–29 (2019)
Nei, Y.-w., Crampton, K.T., Berden, G., Oomens, J., Rodgers, M.T.: Infrared multiple photon dissociation action spectroscopy of deprotonated RNA mononucleotides: Gas-Phase conformations and energetics. J. Phys. Chem. A. 117, 10634–10649 (2013)
Nei, Y.-w., Hallowita, N., Steill, J.D., Oomens, J., Rodgers, M.T.: Infrared multiple photon dissociation action spectroscopy of deprotonated DNA mononucleotides: Gas-Phase conformations and energetics. J. Phys. Chem. A. 117, 1319–1335 (2013)
Wu, R.R., He, C.C., Hamlow, L.A., Nei, Y.-w., Berden, G., Oomens, J., Rodgers, M.T.: Protonation induces base rotation of purine nucleotides pdGuo and pGuo. Phys. Chem. Chem. Phys. 18, 15081–15090 (2016)
Wu, R.R., He, C.C., Hamlow, L.A., Nei, Y.-w., Berden, G., Oomens, J., Rodgers, M.T.: N3 protonation induces base rotation of 2′-deoxyadenosine-5′-monophosphate and adenosine-5′-monophosphate. J. Phys. Chem. B. 120, 4616–4624 (2016)
Wu, R.R., Hamlow, L.A., He, C.C., Nei, Y.-w., Berden, G., Oomens, J., Rodgers, M.T.: The intrinsic basicity of the phosphate backbone exceeds that of uracil and thymine residues: Protonation of the phosphate moiety is preferred over the nucleobase for pdThd and pUrd. Phys. Chem. Chem. Phys. 19, 30351–30361 (2017)
Wu, R.R., Hamlow, L.A., He, C.C., Nei, Y.-w., Berden, G., Oomens, J., Rodgers, M.T.: N3 and O2 protonated conformers of the cytosine mononucleotides coexist in the gas phase. J. Am. Soc. Mass Spectrom. 28, 1638–1646 (2017)
Pedersen, S.O., Stochkel, K., Byskov, C.S., Baggesen, L.M., Nielsen, S.B.: Gas-phase spectroscopy of protonated adenine, adenosine 5′-monophosphate and monohydrated ions. Phys. Chem. Chem. Phys. 15, 19748–19752 (2013)
Chiavarino, B., Crestoni, M.E., Fornarini, S., Lanucara, F., Lemaire, J., Maitre, P., Scuderi, D.: Infrared spectroscopy of isolated nucleotides 1. The cyclic 3′,5′-adenosine monophosphate anion. Int. J. Mass Spectrom. 270, 111–117 (2008)
Lanucara, F., Crestoni, M.E., Chiavarino, B., Fornarini, S., Hernandez, O., Scuderi, D., Maitre, P.: Infrared spectroscopy of nucleotides in the gas phase 2. The protonated cyclic 3′,5′-adenosine monophosphate. RSC Adv. 3, 12711–12720 (2013)
Ligare, M.R., Rijs, A.M., Berden, G., Kabelac, M., Nachtigallova, D., Oomens, J., de Vries, M.S.: Resonant infrared multiple photon dissociation spectroscopy of anionic nucleotide monophosphate clusters. J. Phys. Chem. B. 119, 7894–7901 (2015)
Salpin, J.Y., MacAleese, L., Chirot, F., Dugourd, P.: Structure of the Pb2+-deprotonated dGMP complex in the gas phase: A combined MS-MS/IRMPD spectroscopy/ion mobility study. Phys. Chem. Chem. Phys. 16, 14127–14138 (2014)
Gidden, J., Bowers, M.T.: Gas-phase conformations of deprotonated and protonated mononucleotides determined by ion mobility and theoretical modeling. J. Phys. Chem. B. 107, 12829–12837 (2003)
Dunin-Horkawicz, S., Czerwoniec, A., Gajda, M.J., Feder, M., Grosjean, H., Bujnicki, J.M.: Modomics: A database of RNA modification pathways. Nucleic Acids Res. 34, D145–D149 (2006)
Czerwoniec, A., Dunin-Horkawicz, S., Purta, E., Kaminska, K.H., Kasprzak, J.M., Bujnicki, J.M., Grosjean, H., Rother, K.: Modomics: A database of RNA modification pathways. 2008 update. Nucleic Acids Res. 37, D118–D121 (2009)
Machnicka, M.A., Milanowska, K., Oglou, O.O., Purta, E., Kurkowska, M., Olchowik, A., Januszewski, W., Kalinowski, S., Dunin-Horkawicz, S., Rother, K.M., Helm, M., Bujnicki, J.M., Grosjean, H.: Modomics: A database of RNA modification pathways-2013 update. Nucleic Acids Res. 41, D262–D267 (2013)
Limbach, P.A., Crain, P.F., McCloskey, J.A.: Summary: The modified nucleosides of RNA. Nucleic Acids Res. 22, 2183–2196 (1994)
Cantara, W.A., Crain, P.F., Rozenski, J., McCloskey, J.A., Harris, K.A., Zhang, X.N., Vendeix, F.A.P., Fabris, D., Agris, P.F.: The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 39, D195–D201 (2011)
Bjork, G.R., Durand, J.M.B., Hagervall, T.G., Leipuviene, R., Lundgren, H.K., Nilsson, K., Chen, P., Qian, Q., Urbonavicius, J.: Transfer RNA modification: Influence on translational frameshifting and metabolism. FEBS Lett. 452, 47–51 (1999)
Urbonavicius, J., Qian, O., Durand, J.M.B., Hagervall, T.G., Bjork, G.R.: Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 20, 4863–4873 (2001)
Yarian, C., Townsend, H., Czestkowski, W., Sochacka, E., Malkiewicz, A.J., Guenther, R., Miskiewicz, A., Agris, P.F.: Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 277, 16391–16395 (2002)
Agris, P.F., Vendeix, F.A.P., Graham, W.D.: tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 366, 1–13 (2007)
Kawai, G., Yamamoto, Y., Kamimura, T., Masegi, T., Sekine, M., Hata, T., Iimori, T., Watanabe, T., Miyazawa, T., Yokoyama, S.: Conformational rigidity of specific pyrimidine residues in transfer-RNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry-US. 31, 1040–1046 (1992)
Yokoyama, S., Watanabe, T., Murao, K., Ishikura, H., Yamaizumi, Z., Nishimura, S., Miyazawa, T.: Molecular mechanism of codon recognition by transfer-RNA species with modified uridine in the 1st position of the anticodon. Proc. Natl. Acad. Sci. USA. 82, 4905–4909 (1985)
Sprinzl, M., Hartmann, T., Weber, J., Blank, J., Zeidler, R.: Compilation of transfer-RNA sequences and sequences of transfer genes. Nucleic Acids Res. 17, R1–R172 (1989)
Ladner, J.E., Jack, A., Robertus, J.D., Brown, R.S., Rhodes, D., Clark, B.F.C., Klug, A.: Structure of yeast phenylalanine transfer-RNA at 2.5 Å resolution. Proc. Natl. Acad. Sci. USA. 72, 4414–4418 (1975)
Gross, H.J., Simsek, M., Raba, M., Limburg, K., Heckman, J., Rajbhandary, U.L.: 2'-O-methyl ribothymidine: A component of rabbit liver lysine transfer-RNA. Nucleic Acids Res. 1, 35–43 (1974)
Lubini, P., Zurcher, W., Egli, M.: Stabilizing effects of the RNA 2′-substituent: Crystal structure of an oligodeoxynucleotide duplex containing 2'-O-methylated adenosines. Chem. Biol. 1, 39–45 (1994)
Prusiner, P., Sundaralingam, M.: Molecular and crystal structure of the modified nucleoside 2'-O-methyladenosine. A novel 2′-exo-3′-endo 2T3 sugar pucker. Acta Crystallogr. B. 32, 161–169 (1976)
Chow, S., Wen, K., Sanghvi, Y.S., Theodorakis, E.A.: Novel synthesis of 2'-O-methylguanosine. Bioorg. Med. Chem. Lett. 13, 1631–1634 (2003)
Lee, C.H., Tinoco, I., Jr.: Studies of the conformation of modified dinucleoside phosphates containing 1,N6-ethenoadenosine and 2'-O-methylcytidine by 360-MHz 1H nuclear magnetic resonance spectroscopy. Investigation of the solution conformations of dinucleoside phosphates. Biochemistry-US. 16, 5403–5414 (1977)
Barbe, S., Le Bret, M.: Effect of a water molecule on the sugar puckering of uridine, 2′-deoxyuridine, and 2'-O-methyl uridine inserted in duplexes. J. Phys. Chem. A. 112, 989–999 (2008)
Yathindra, N., Sundaralingam, M.: Effect of O(2′)-methylation on the stability of polynucleotide helices. Biopolymers. 18, 2721–2731 (1979)
Hingerty, B., Bond, P.J., Langridge, R., Rottman, F.: Conformation of 2'-O-methyl cytidine, a modified furanose component of ribonucleic acids. Biochem. Biophys. Res. Commun. 61, 875–881 (1974)
Cheng, D.M., Sarma, R.H.: Nuclear magnetic resonance study of the impact of ribose 2'-O-methylation on the aqueous solution conformation of cytidylyl-(3′->5′)-cytidine. Biopolymers. 16, 1687–1711 (1977)
Kawai, G., Ue, H., Yasuda, M., Sakamoto, K., Hashizume, T., McCloskey, J. A., Miyazawa, T., Yokoyama, S.: Relation between functions and conformational characteristics of modified nucleosides found in tRNAs. Nucleic Acids Symp. Ser. 49–50 (1991).
Masaki, Y., Miyasaka, R., Ohkubo, A., Seio, K., Sekine, M.: Linear relationship between deformability and thermal stability of 2'-O-modified RNA hetero duplexes. J. Phys. Chem. B. 114, 2517–2524 (2010)
Majlessi, M., Nelson, N.C., Becker, M.M.: Advantages of 2'-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 26, 2224–2229 (1998)
Lesnik, E.A., Guinosso, C.J., Kawasaki, A.M., Sasmor, H., Zounes, M., Cummins, L.L., Ecker, D.J., Cook, P.D., Freier, S.M.: Oligodeoxynucleotides containing 2'-O-modified adenosine - synthesis and effects on stability of DNA-RNA duplexes. Biochemistry-US. 32, 7832–7838 (1993)
Egli, M.: Structural aspects of nucleic acid analogs and antisense oligonucleotides. Angew. Chem. Int. Ed. 35, 1895–1910 (1996)
Lesnik, E.A., Freier, S.M.: What affects the effect of 2′-alkoxy modifications? 1. Stabilization effect of 2′-methoxy substitutions in uniformly modified DNA oligonucleotides. Biochemistry-US. 37, 6991–6997 (1998)
Davis, D. R.: Biophysical and conformational properties of modified nucleosides in RNA (nuclear magnetic resonance studies). In: Grosjean, H., Benne, R. (eds.) Modification and editing of RNA, chap. 5, pp. 85–102. ASM Press, Washington, DC (1998)
Valle, J.J., Eyler, J.R., Oomens, J., Moore, D.T., van der Meer, A.F.G., von Helden, G., Meijer, G., Hendrickson, C.L., Marshall, A.G., Blakney, G.T.: Free electron laser-Fourier transform ion cyclotron resonance mass spectrometry facility for obtaining infrared multiphoton dissociation spectra of gaseous ions. Rev. Sci. Instrum. 76, 23103 (2005)
Polfer, N.C., Oomens, J., Moore, D.T., von Helden, G., Meijer, G., Dunbar, R.C.: Infrared spectroscopy of phenylalanine Ag(I) and Zn (II) complexes in the gas phase. J. Am. Chem. Soc. 128, 517–525 (2006)
Polfer, N.C., Oomens, J.: Reaction products in mass spectrometry elucidated with infrared spectroscopy. Phys. Chem. Chem. Phys. 9, 3804–3817 (2007)
Maitre, P., Le Caer, S., Simon, A., Jones, W., Lemaire, J., Mestdagh, H.N., Heninger, M., Mauclaire, G., Boissel, P., Prazeres, R., Glotin, F., Ortega, J.M.: Ultrasensitive spectroscopy of ionic reactive intermediates in the gas phase performed with the first coupling of an IR FEL with an FTICR-MS. Nucl. Instrum. Methods Phys. Res. A. 507, 541–546 (2003)
Stuart, B.H.: Infrared spectroscopy: Fundamentals and applications. In: Ando, D.J. (ed.) Analytical techniques in the sciences. John Wiley & Sons, Ltd, Chichester (2004)
Rijs, A., Oomens, J.: Gas-phase IR spectroscopy and structure of biological molecules. Top. Curr. Chem. (2015).
MacAleese, L., Maitre, P.: Infrared spectroscopy of organometallic ions in the gas phase: from model to real world complexes. Mass Spectrom. Rev. 26, 583–605 (2007)
Lemaire, J., Boissel, P., Heninger, M., Mauclaire, G., Bellec, G., Mestdagh, H., Simon, A., Le Caer, S., Ortega, J.M., Glotin, F., Maitre, P.: Gas phase infrared spectroscopy of selectively prepared ions. Phys. Rev. Lett. 89, 273002-3 (2002)
Bakker, J.M., Besson, T., Lemaire, J., Scuderi, D., Maitre, P.: Gas-phase structure of a π-allyl-palladium complex: efficient infrared spectroscopy in a 7 T Fourier transform mass spectrometer. J. Phys. Chem. A. 111, 13415–13424 (2007)
Prazeres, R., Glotin, F., Insa, C., Jaroszynski, D.A., Ortega, J.M.: Two-colour operation of a free-electron laser and applications in the mid-infrared. Eur. Phys. J. D. 3, 87–93 (1998)
Bakker, J.M., Sinha, R.K., Besson, T., Brugnara, M., Tosi, P., Salpin, J.Y., Maitre, P.: Tautomerism of uracil probed via infrared spectroscopy of singly hydrated protonated uracil. J. Phys. Chem. A. 112, 12393–12400 (2008)
Schindler, B., Joshi, J., Allouche, A.R., Simon, D., Chambert, S., Brites, V., Gaigeot, M.P., Compagnon, I.: Distinguishing isobaric phosphated and sulfated carbohydrates by coupling of mass spectrometry with gas phase vibrational spectroscopy. Phys. Chem. Chem. Phys. 16, 22131–22138 (2014)
Schindler, B., Renois-Predelus, G., Bagdadi, N., Melizi, S., Barnes, L., Chambert, S., Allouche, A.R., Compagnon, I.: MS/IR, a new MS-based hyphenated method for analysis of hexuronic acid epimers in glycosaminoglycans. Glycoconj. J. 34, 421–425 (2017)
Schindler, B., Barnes, L., Gray, C.J., Chambert, S., Flitsch, S.L., Oomens, J., Daniel, R., Allouche, A.R., Compagnon, I.: IRMPD spectroscopy sheds new (infrared) light on the sulfate pattern of carbohydrates. J. Phys. Chem. A. 121, 2114–2120 (2017)
Bertsimas, D., Tsitsiklis, J.: Simulated annealing. Stat. Sci. 8, 10–15 (1993)
Wolinski, K., Hinton, J. F., Wishart, D. S., Sykes, B. D., Richards, F. M., Pastone, A., Saudek, V., Ellis, P. D., Maciel, G. E., McIver, J. W., Jr, Blizzard, A. C., Santry, D. P., Pople, J. A., Ostlund, N. S., Ducasse, L., Hoarau, J., Pesquer, M., Kondo, M., Ando, I., Chujo, R., Nishioka, A., Vauthier, E. C., Odiot, S., Tonnard, F., Baker, J. D., Zerner, M. C., Beveridge, D. V., Anderson, W. P., Cundari, T. R., Bingham, R. C., Dewar, M. J. S., Lo, D. H., Li, J., Mello, P. C., Jug, K., Thiel, W., Zoebisch, E. G., Healy, E. F., Stewart, J. J. P., Fraser, M., Hayes, D. M.: HyperChem(TM) Professional 8.0, Hypercube, Inc., Gainesville, FL (2004)
Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Scalmani, G., Barone, V., Mennucci, B., Petersson, G. A., Nakatsuji, H., Caricato, M., Li, X., Hratchian, H. P., Izmaylov, A. F., Bloino, J., Zheng, G., Sonnenberg, J. L., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Vreven, T., Montgomery, J.A., Jr., Peralta, J. E., Ogliaro, F., Bearpark, M. J., Heyd, J., Brothers, E. N., Kudin, K. N., Staroverov, V. N., Kobayashi, R., Normand, J., Raghavachari, K., Rendell, A. P., Burant, J. C., Iyengar, S. S., Tomasi, J., Cossi, M., Rega, N., Millam, N. J., Klene, M., Knox, J. E., Cross, J. B., Bakken, V., Adamo, C., Jaramillo, J., Gomperts, R., Stratmann, R. E., Yazyev, O., Austin, A. J., Cammi, R., Pomelli, C., Ochterski, J. W., Martin, R. L., Morokuma, K., Zakrzewski, V. G., Voth, G. A., Salvador, P., Dannenberg, J. J., Dapprich, S., Daniels, A. D., Farkas, Ö., Foresman, J. B., Ortiz, J. V., Cioslowski, J., Fox, D. J.: Gaussian 09, Revision A.02, Gaussian, Inc., Wallingford, CT, USA (2016).
Hunter, E.P., Lias, S.G.: Proton affinity evaluation. In: Linstrom, P.J., Mallard, W.G. (eds.) NIST chemistry webbook, NIST standard reference database number 69. National Institute of Standards and Technology, Gaithersburg (2018)
Wu, R.R., Rodgers, M.T.: Tautomerization lowers the activation barriers for N-glycosidic bond cleavage of protonated uridine and 2′-deoxyuridine. Phys. Chem. Chem. Phys. 18, 24451–24459 (2016)
Wu, R.R.: Studies Toward A Detailed Understanding Of The Intrinsic Properties Of Nucleic Acid Constituents: Gas-Phase Conformations, Energetics, and Glycosidic Bond Stability (2016). Wayne State University Dissertations. 1496.
Wu, R.R., Rodgers, M.T.: O2 protonation controls threshold behavior for N-glycosidic bond cleavage of protonated cytosine nucleosides. J. Phys. Chem. B. 120, 4803–4811 (2016)
Altona, C., Sundaralingam, M.: Conformational-analysis of sugar ring in nucleosides and nucleotides - New description using concept of pseudorotation. J. Am. Chem. Soc. 94, 8205–8212 (1972)
Saenger, W. Principles of Nucleic Acid Structure, Springer-Verlag, New York pp. 51–104 (1984).
Moore, D.T., Oomens, J., van der Meer, L., von Helden, G., Meijer, G., Valle, J., Marshall, A.G., Eyler, J.R.: Probing the vibrations of shared, OH+O-bound protons in the gas phase. ChemPhysChem. 5, 740–743 (2004)
Heine, N., Fagiani, M.R., Rossi, M., Wende, T., Berden, G., Blum, V., Asmis, K.R.: Isomer-selective detection of hydrogen-bond vibrations in the protonated water hexamer. J. Am. Chem. Soc. 135, 8266–8273 (2013)
Cooper, T.E., O'Brien, J.T., Williams, E.R., Armentrout, P.B.: Zn2+ has a primary hydration sphere of five: IR action spectroscopy and theoretical studies of hydrated Zn2+ complexes in the gas phase. J. Phys. Chem. A. 114, 12646–12655 (2010)
Contreras, C.S., Polfer, N.C., Oomens, J., Steill, J.D., Bendiak, B., Eyler, J.R.: On the path to glycan conformer identification: Gas-phase study of the anomers of methyl glycosides of N-acetyl-D-glucosamine and N-acetyl-D-galactosamine. Int. J. Mass Spectrom. 330, 285–294 (2012)
Santini, G.P.H., Pakleza, C., Auffinger, P., Moriou, C., Favre, A., Clivio, P., Cognet, J.A.H.: Dinucleotide tpt and its 2'-O-Me analogue possess different backbone conformations and flexibilities but similar stacked geometries. J. Phys. Chem. B. 111, 9400–9409 (2007)
Acknowledgements
This work is financially supported by the National Science Foundation, under Grants OISE-0730072 and OISE-1357787 (for international travel expenses), DBI-0922819 (for the Bruker amaZon ETD QITMS employed in this work), and CHE-1709789 and CHE-1664618 (for other research costs). C.C.H., L.A.H., Y.Z., and Y.-w.N. are supported by the Wayne State University Thomas C. Rumble Graduate Fellowships and Summer Dissertation Fellowships. We thank Wayne State University C&IT for excellent computational resources and support. The skillful assistance of the CLIO staff is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
ESM 1
(PDF 19328 kb)
Rights and permissions
About this article
Cite this article
He, C.C., Hamlow, L.A., Zhu, Y. et al. Structural and Energetic Effects of O2′-Ribose Methylation of Protonated Pyrimidine Nucleosides. J. Am. Soc. Mass Spectrom. 30, 2318–2334 (2019). https://doi.org/10.1007/s13361-019-02300-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-019-02300-9
Keywords
- Cytidine (Cyd)
- Cytosine (Cyt)
- Density functional theory (DFT)
- Electronic structure calculations
- Electrospray ionization (ESI)
- Energy-resolved collision-induced dissociation (ER-CID)
- Fourier transform ion cyclotron resonance mass spectrometer (FT-ICR MS)
- Gas-phase conformation
- Glycosidic bond stability
- Hydrogen-bonding interactions
- Hydrogen-stretching region
- Infrared multiple photon dissociation (IRMPD) action spectroscopy
- IR fingerprint region
- IRMPD spectrum
- IR spectrum
- 5-Methyluridine
- Nucleobase
- Nucleobase orientation
- Nucleoside
- Nucleoside modification
- 2′-O-methylation
- 2′-O-methylcytidine (Cydm)
- 2′-O-methyl-5-methyluridine (Thdm)
- 2′-O-methyluridine (Urdm)
- Protonation
- Pyrimidine nucleosides
- Quadrupole ion trap mass spectrometer (QIT MS)
- Simulated annealing
- Sugar puckering
- Survival yield analysis
- Tandem mass spectrometry
- Thymidine (Thd)
- Thymine (Thy)
- Uracil (Ura)
- Uridine (Urd)