Abstract
Study on expression of genes for the traits associated with hypoxia tolerance during the germination demands robust choice of reference genes for transcript data normalization and gene validation through real-time quantitative polymerase chain reaction (RT-qPCR). However, reliability and stability of reference genes across different rice germplasms under hypoxic condition have not been accessed yet. Stability performance of reference genes such as eukaryotic elongation factor 1 α (eEF1α), ubiquitin 10 (UBQ10), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), 18S ribosomal RNA (18SrRNA), 25S ribosomal RNA (25SrRNA), β-tublin (β-TUB), actin11 (ACT11), ubiquitin C (UBC), eukaryotic elongation factor 4 α (eIF4α), and ubiquitin5 (UBQ5) was accessed through statistical algorithms like geNorm, NormFinder, Comparative ΔCt method, BestKeeper, and RefFinder in three rice germplasms (KHO, RKB, and IR-64) with varied level of tolerance to hypoxic condition during germination. Among all genes used, OsGAPDH was found to be the most suitable reference gene under hypoxic condition. The performance of the highest-ranking reference gene (OsGAPDH) in terms of stability based on statistical algorithms was further validated for its reliability and stability through RT-qPCR with hypoxia-induced target gene OsTTP7. The identified stable housekeeping gene could be used as internal control for gene expression analysis in rice under hypoxia.
Similar content being viewed by others
Introduction
Devastation due to flooding during sowing and transplanting of rice leads to severe crop loss (Jackson and Ram 2003; Setter et al. 1997). Several rice genotypes exhibiting a different level of tolerance to hypoxic condition during germination were identified through germplasm screening (Septiningsih et al. 2013). A tremendous advancement in breeding for hypoxic germination-tolerant rice varieties supplemented with high throughput tools for genomic studies for quantitative assessment of genes at transcript level provides a greater opportunity to understand the genetic and molecular basis of hypoxia tolerance. The signal cascade emanating from the perception of stimuli led to activation of hypoxia-responsive genes through various transcription factors, either at the transcriptional or post-transcriptional level results in re-programming of adaptive response at metabolic and physiological level (Sasidharan and Mustroph 2011). Changes in signaling and transcriptional regulation are species-specific (Mustroph et al. 2010). For these instances, quantitative real-time PCR (RT-qPCR), an analytical technique known for high sensitivity and reproducibility, has been used for the gene expression analysis (Chi et al. 2012). The first advantage of RT-qPCR is that it accurately quantifies gene expression without any follow-up intervention, thus reducing the experimental error (Nolan et al. 2006). Reportedly, the RT-qPCR has been used to examine the pattern of gene expression affected by flooding in Arabidopsis(Liu et al. 2005), rice (Kretzschmar et al. 2015; Lasanthi-Kudahettige et al. 2007), maize (Zou et al. 2010), populous (Kreuzwieser et al. 2009), and cotton (Christianson et al. 2010). RNA quality and integrity, variation in the RNA initial amount, and efficiency of complementary DNA (cDNA) synthesis may affect the PCR performance. A typical well-known strategy to correct for analytical variation is to normalize the expression of the target gene against stable endogenous gene(s). In order to qualify for an endogenous control in RT-qPCR, a gene should display stable expression in the plant, irrespective of the developmental and environmental condition (Bo et al. 2008). Yet, to avoid any inconsistency in gene expression of the target genes due to suboptimal performance of endogenous control, it is necessary to validate the candidate reference genes for a set of treatment and different sample types (Xu et al. 2011). The statistical tools such as geNorm, Norm Finder, and BestKeeper (Setter et al. 1997; Christianson et al. 2010; Xu et al. 2015) have been used for determining the stability analysis of the reference genes. RefFinder is another statistical tool that has been used for grouping all the above-mentioned algorithms (Mallona et al. 2010). Housekeeping genes such as OsGAPDH and ACT-B have been frequently used in the gene expression studies (Thellin et al. 1999; Bustin et al. 2009). However, reliability and stability of reference genes in rice germplasms under hypoxic condition during germination have not been reported elsewhere. The goal of the present investigation was to identify the best stable candidate reference gene for gene expression studies in rice under hypoxia. We applied open-access software such as RefFinder (Mallona et al. 2010) for the stability analysis and ranking of ten housekeeping genes to narrow-down on the most appropriate gene for normalization under hypoxia. Our findings are likely to reveal that the highest-ranked reference gene obtained using statistical algorithms also performed with greater stability when validated against known hypoxia-induced gene like OsTTP7 during rice seed germination. The identified reference gene can therefore be used for normalization of candidate gene in rice germination studies under hypoxia.
Materials and methods
Plant material selection and hypoxic treatment
Two AG (anaerobic germination)-tolerant rice (Oryzasativa L.) germplasms, Khao Hlan On (KHO, obtained from IRRI, Philippines), Rangadhar Kekua Bao (Deepwater rice ecotype from Assam, India), and an AG-susceptible variety IR-64 were used for the gene expression studies under hypoxic condition. Seeds of hypoxia-susceptible and tolerant germplasms were sown in bottles containing sterile water at a depth of 10 cm and maintained at 30 °C in dark in an incubator for up to 96 h (O2 concentration was 2–2.5% as indicated by DO meter at seed depth). These seeds were grown in petri dish containing water-soaked filter to check the viability of the seeds. For stability assessment of housekeeping genes in hypoxic-tolerant and susceptible germplasms, samples for RNA isolation were collected at 24 h, 48 h, and 96 h after sowing and stored immediately in liquid nitrogen until further processing. As OsTPP7 gene is transcriptionally upregulated under hypoxic condition in tolerant rice, which takes part in the conversion of trehalose-6-phosphate to trehalose (Kretzschmar et al. 2015), this gene was used as a target gene to validate reference genes for the gene expression studies during germination under hypoxia. Each experiment was done taking 3 biological replications. Sampling analysis was done with three technical replicates.
Designing of primers
The stability of the qPCR was analyzed using 10 candidate reference genes. All genes used in the present study were present in single copy in rice genome, and the primers were used based on sequence information previously cited in rice reference gene validation studies (Jain et al., 2006). The expected amplicon sizes for reference genes used, namely eukaryotic elongation factor 1 α (eEF1α), ubiquitin 10 (UBQ10), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), 18S ribosomal RNA (18SrRNA), 25S ribosomal RNA (25SrRNA), β-tubulin (β-TUB), actin11 (ACT11), ubiquitin C (UBC), eukaryotic elongation factor 4 α (eIF4α), and ubiquitin5 (UBQ5) were obtained from the published report (Jain et al. 2006). Primer pairs were tested for Tm, stability, GC content, and hairpin loop by using oligo-dT analyzer. The primer sequences and amplicon characteristics of the selected reference genes are listed in Table 1. The specificity of the amplicon was verified by dissociation curve analysis (60 to 95 °C) after 40 cycles of PCR and by agarose gel electrophoresis.
Isolation of total RNA and cDNA synthesis
Total RNA was isolated from seeds collected at different time points using Pure-link reagent (Invitrogen, USA, cat no. 12322-012) following manufacturer’s instructions. Purification of RNA was carried out using DNase I (Sigma Aldrich, cat no. AMPD1) and the RNA concentration was determined using Nano-drop 2000 (Thermo Fisher Scientific Wilminton D, USA). The integrity of the RNA samples was analyzed using 1.5% agarose gel electrophoresis stained with ethidium bromide. One microgram (1 μg) of RNA was used for cDNA synthesis using the Prime Script RT reagent Kit (Takara, Japan cat no. RR047A) following the manufacturer’s guidelines.
Quantitative real-time PCR assay
RT-qPCR reactions were performed on an Applied Biosystem Step One Plus System using SYBR Premix Ex Taq (Takara, Japan). PCR conditions were optimized based on the primer efficiency. Briefly, a 10-μL reaction mixture consisted of 5 μL of SYBR Premix Ex Taq,0.2 μL of ROX dye, 2 μL of cDNA (50 ng/1:20 dilution), and 1.8 μL of Nuclease-Free Water (Ambion, USA cat no AM9930) using three different primer concentrations, along with a negative control (without cDNA). Thermal cycle used for all RT-qPCR reactions was 30 s at 95 °C followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. After 40 cycles the specificity of the amplicon was analyzed through the dissociation curve profile and agarose gel electrophoresis (2%). Each reaction was performed with three biological and three technical replications.
Selection of candidate reference genes based on ranking
The primer efficiency and specificity of all reference genes were evaluated prior to the expression profile of housekeeping genes during RT-qPCR. Primer specificity was evaluated with the help of dissociation curve. For determination of the best candidate reference gene pair, the following statistical algorithms were used: NormFinder (Andersen et al. 2004), GeNorm (Vandesompele et al. 2002), Best Keeper (Pfaffl et al. 2004), and ΔCt method (Silver et al. 2006), and RefFinder (Mallona et al. 2010), web-based software. A set of 4-fold dilutions (1:20; 1:40; 1:80; 1:160) of cDNA from RKB, KHO, and IR-64were used to make the standard curves; thus, the PCR efficiency (E) and correlation coefficient (R2) were determined for each gene using the linear regression model. The PCR efficiency was calculated as E = (10–1/slope) − 1.
Data analyses
BestKeeper algorithm determined the optimal housekeeping gene employing the pairwise correlation analysis based on the standard deviation (±CP) of all the pairs of tested reference genes (Reena et al. 2009). GeNorm algorithm, which is a stepwise exclusion method based on average expression stability (M) value, provided a pair of two housekeeping genes having equal expression levels (Vandesompele et al. 2002). The NormFinder, an Excel-based algorithm, determined the expression stability value of the candidate gene and identified the stable housekeeping genes based on intra- and inter-group variations among the tested genes (Andersen et al. 2004). For geNorm and NormFinder, the raw Cq values were converted into relative quantity values using the formula 2 − ΔCq. Finally, RefFinder was employed for combining all the algorithms used to find the best suitable reference gene (Mallona et al. 2010). All the measurements were carried out in triplicate, and for each primer set, reaction efficiency estimates were derived from standard curves that were generated using serial dilutions of a cDNA pool of all the rice samples. These were then used by qBase to transform the Ct values to relative quantities for analysis with geNorm 3.4 software. After normalization using the geometric mean of the most stable reference genes, the normalized OsTPP7 expression level ratios were standardized to minimize inter-experimental variation. For the purpose, the normalized expression levels were converted into logarithmic values, divided by their standard deviations and multiplied by the mean standard deviation of all the experiments to calculate the mean standardized expression level per rice sample along with their respective 95% confidence intervals. Finally, all the values were linearized using a power function and plotted in a graph. Differential gene expression was considered to be significant when the 95% confidence interval of the mean expression levels did not overlap (equivalent to P < 0.05).
Results
Selection of housekeeping genes for the stable expression during hypoxia
The qPCR amplification efficiency (E) ranged from 90.30 to 104.10%, with a correlation coefficient (R2) ranging from 0.982 to 0.998. The mean Ct values of 10 candidate genes in all samples varied from 10.25468 to 34.37549 (Table 1). For each primer, the dissociation curve displayed a unique peak of fluorescence, indicating that a single fragment was amplified during qPCR amplification for samples of all treatments (Fig. S1). The cycle threshold (Ct) values of 10 reference genes in different germplasms under three different treatments were used to compare the gene expression pattern. Comparative analyses of all the genes tested revealed a wide range of expression variances of the Ct values under all three treatments with the highest variation observed in Os25SRNA and Os18SRNA and lower variation in OsGAPDH, OseEF1α, and OseIF4α. Even though the variation analysis based on Ct values revealed some of the genes with relatively less variation, a comparison with other statistical algorithms was necessary for determining the most stable gene(s) for normalizing target gene expression.
Analysis of the housekeeping genes based on a geNorm algorithm
GeNorm analysis ranked the target reference gene according to their average expression stability (M value) using the Ct value of all samples. Samples with lowest M value were considered as the most stable and vice versa. The M value of housekeeping genes ranged from 0.868 to 3.575 (Min-Max). OseIF4α/Osβ-TUB showed the highest stability (M value of 0.868) and Os25SrRNA (M value of 3.575) was the least stable gene among all reference genes tested (Fig. 1).
Analysis of the housekeeping genes based on a NormFinder algorithm
The NormFinder analysis, which is based on intra- and inter-group variations, was employed for the estimation of stability values. Following this approach, OsEF1α (stability value, 0.569) was identified as the most stable gene followed by OsGAPDH (stability value, 0.685), OsACT 11 (stability value, 4.563), and Os25SrRNA (stability value 6.236) (Fig. 2).
Expression stability analysis of each candidate reference gene among different anaerobic treated tissue samples using NormFinder algorithm. Lower stability value indicates higher stability of the housekeeping gene and vice versa. The direction of the arrows indicates the most and least stable housekeeping genes
Analysis of the housekeeping genes based on the comparative ΔCт method
The stability of the housekeeping genes was determined by using comparative ΔCт methods based on standard deviation (SD). Higher SD value correlated to the low stability of the housekeeping genes and vice versa. The OsGAPDH with SD value of 2.63 was found to be the most stable housekeeping gene while Os25SrRNA having SD value of 5.3 was the least stable (Fig. 3).
Expression stability analysis of each candidate reference gene among different anaerobic-treated tissue samples using comparative ΔCт method. Lower stability value indicates higher stability of the housekeeping gene and vice versa. The direction of the arrows indicates the most and least stable housekeeping genes
Analysis of the housekeeping genes based on the BestKeeper algorithm
The stability of the housekeeping genes was determined by BestKeeper based on the extent of standard deviation (SD [±CP) with higher SD value corresponding to the low stability of the housekeeping genes and vice versa. The descriptive statistics of all the ten housekeeping genes computed by this algorithm (Fig. 4) led to the identification of six housekeeping genes (OsUBC,OsACT11, Osβ-TUB, OsGAPDH, OseIF4α, andOsEF1α) which were consistent and stable. On the other hand, the other 4 genes, namely Os25SrRNA (SD, 5.39), Os18SrRNA (SD, 4.31), OsUBQ5 (SD, 3.48), and OsUBQ10 (SD, 3.39) were found to be inconsistent and less stable. The coefficient of variation (CV %) of all housekeeping genes ranged from 5.37% for OsUBC to 27.06% for Os25SrRNA, suggesting different levels of variation in candidate housekeeping genes (Table S1).
Expression stability analysis of each candidate reference gene among different anaerobically treated tissue samples using BestKeeper. The lower standard deviation (SD) [±CP] value indicates the higher the stability of the gene and vice versa. The direction of the arrows indicates the most and least stable reference genes used in the study
Comparative expression stability analysis based on RefFinder algorithm
As expression stability analysis of target genes accessed using proven statistical algorithm showed inconsistency in terms of their stability, a comparative gene expression stability and ranking of genes based on their Ct value was performed through RefFinder. Based on the stability value, four housekeeping genes namely OsGAPDH (geo mean 2.21), OseIF4α (geo mean 2.59), OsEF1α (geo mean 2.63), and Osβ-TUB (geo mean 2.63) were found to be stable across different treatments, while UBQ5 and 25SrRNA with a Geo Mean of 7.74 and 10.0, respectively, were found to be less stable (Fig. 5).Thus, comparative study of all four statistical methods provided a way for ranking the housekeeping genes based on their performance (Table 2). Among all, OsGAPDH was considered to be the most stable housekeeping gene for the gene expression studies under hypoxia in rice during germination and coleoptile elongation.
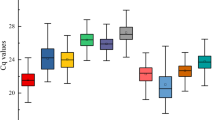
Ct variation and expression stability analysis of each candidate reference gene among different tissue samples using RefFinder. a Boxplot depicting absolute Ct values, which were calculated using GenEx program. Lower and upper boxes indicate the 25th and 75th percentile, respectively, and median is depicted by the line. b Gene expression stability graph using RefFinder algorithm based on stability values of all algorithm. The direction of the arrows indicates the most and least stable housekeeping genes
Validation of identified stable reference genes
Analyses of data for different hours of treatment (24 h, 48 h, and 96 h) under the hypoxic condition of OsTPP7 gene revealed differences in the pattern of expression when normalized with the most stable and the least stable housekeeping genes (25SrRNA). Relative quantification (RQ) of OsTPP7 gene varied according to the internal control (stable and least stable) used during normalization of the target gene.
Analysis of dataset for three genotypes (KHO, RKB, and IR-64) at 24 h (hypoxic) time point revealed significant expression difference when normalized against the most stable and the least stable housekeeping genes. RQ of target candidate gene, OsTPP7, was higher when normalized against the stable internal control genes, such as OSGAPDH (2.70-fold in RKB and 3.42-fold in KHO), OsEF1α (3.53-fold in RKB and 3.21-fold in KHO) and eIF4α (4.14-fold in RKB and 3.65-fold in KHO) compared to more commonly used housekeeping gene OsACT 11 (2.01-fold in RKB and 2.51-fold in KHO).
Likewise, analysis of dataset at 48-h hypoxic time point also revealed significant variation in expression level of the target candidate gene (OsTPP7). Fold change in expression of OsTPP7 represented by RQ value was higher when normalized against the stable internal control genes, such as OsGAPDH (0.88-fold for IR64, 4.80-fold in RKB, and 4.15-fold-in KHO), OsEF1α (0.90-fold in IR-64, 4.70-fold in RKB, and 3.83-fold in KHO) and OseIF4α (1.0-fold in IR64, 5.35-fold in RKB, and 3.34-fold in KHO) compared to the commonly used housekeeping gene OsACT 11 (0.89-fold in OsACT 11, 2.76-fold in RKB, and 2.53-fold in KHO). However, the expression of the target gene was more than 3-fold lower in sample normalized with Os25SrRNA at 24 h (0.82-fold in IR64, 0.81-fold in RKB, and 0.73-fold in KHO) compared to corresponding stable internal control (OsGAPDH, OsEF1α, OseIF4α) as well as OsACT 11 normalized samples.
Analysis of dataset at 96-h hypoxia-treated samples showed marked difference in expression levels in susceptible and tolerant genotypes. The fold change of OsTTP7 gene when most stable housekeeping genes such as OsGAPDH were used as internal control (0.58-fold for IR64, 4.77-fold in RKB, and 3.53-fold in KHO), OsEF1α (0.97-fold in IR-64, 3.98-fold in RKB, and 3.93-fold in KHO) and OseIF4α (0.81fold in IR64, 3.15-fold in RKB, and 3.26-fold in KHO) compared to the commonly used housekeeping gene OsACT 11 (0.42-fold in IR64, 2.82-fold in RKB, and 2.29-fold in KHO), the performance of least stable gene Os25SrRNA in terms of expression of target gene in susceptible and tolerant genotypes were similar (0.38-fold in IR-64, 0.76-fold in RKB, 2.89-fold in KHO) (Fig. 6). This finding shows that the stable genes exhibit similar levels of gene expression highlighting the significance of the choice of internal controls under hypoxia for expression profiling of candidate genes.
Normalization of hypoxia-inducible target gene OsTPP7 in tolerant (KHO, RKB) and susceptible (IR-64) cultivar. The relative quantification (RQ) value of OsTPP7 was obtained by normalization with housekeeping genes such as (i) GAPDH, (ii) eEF4α, (iii) eEF1α, (iv) actin, and (v) 25SrRNA gene using susceptible IR-64 at 24 HAG as treatment control
Discussion
Tolerance of rice crop to flooding through enhanced germination and early seedling growth is a prerequisite for successful cultivation of rice in a region where flooding is a recurrent event. Although rice could tolerate flooding, its germination is limited to coleoptile elongation as root and primary leaf fail to develop in susceptible genotypes (Dumbala et al. 2013).
As evident from previous studies, hypoxic germination is a complex process and performance of a tolerant genotype could vary depending on the level of oxygen deprivation (Xie et al. 2011). The process of cell division and elongation is known to occur under hypoxia. However, the ability of cells to divide under the hypoxic condition is determined by the period of submergence, which is around 48 h (Selvey et al. 2005), as beyond which point there would be energy bottlenecks due to lack of oxygen (Ohl et al. 2005). In order to understand the molecular basis of the response of rice to an abiotic stimulus such as hypoxic condition during germination, studies with qPCR have been widely conducted for characterizing gene expression patterns. Normalization of target gene expression via stable endogenous housekeeping genes is a prerequisite to minimize error in relative quantification as the use of inconsistent housekeeping genes may lead to erroneous results leading to misinterpretation of data (Selvey et al. 2005; Ohl et al. 2005). Besides, for accurate quantification of target gene expression, it is essential that expression of reference gene should be independent of genotypes, conditions, and treatments (Isaiah et al. 2016; Maksup et al. 2013; Manoli et al. 2012).Therefore, to perform the accurate candidate gene expression analysis, number of housekeeping genes have been tested and identified in many crops such as rice (Xu et al. 2015; Isaiah et al. 2016; Jain et al. 2006; Maksup et al. 2013), maize (Manoli et al. 2012; Yueai et al. 2014), wheat (Long et al. 2010; Paolacci et al. 2009), soybean (Hu et al. 2009; Le et al. 2012; Ma et al. 2013; Nakayama et al. 2014), peanut (Chi et al. 2012; Reddy et al. 2013), and tea (Ming-Le et al. 2017).
Hypoxic condition prevailing during the time of germination affects global transcript change (Reena et al. 2009). Choice of internal control is of utmost importance when expressions of genes are studied in metabolically active tissues, particularly during early seed germination and coleoptile elongation when there is a rapid change in transcript dynamics (Reena et al. 2017). OsACT11 and OsUBQ have been previously used as reference genes for assessment of transcript perturbation in hypoxic responsive genes in hypoxic-sensitive and tolerant rice (Kretzschmar et al. 2015). We did not come across any published report that systematically analyzes the expression stability of housekeeping genes in rice under hypoxic condition. In order to assess any shortcomings with regard to the stability of housekeeping gene that could affect the performance of target genes for hypoxic germination studies, 10 well-known housekeeping genes were studied on a set of AG-tolerant viz KHO and RKB and susceptible IR-64 rice germplasm through different statistical approaches. Use of different statistical algorithms for validation, normalization, and statistical modeling of selected reference gene has been reported (Ward et al. 2015; Aniko et al. 2004). The BestKeeper was previously used for analyzing descriptive statistics of different reference genes, while geNorm, comparative ΔCт, and NormFinder were used to determine the ranking of genes according to their stability values (Manoli et al. 2012; Ming-Le et al. 2017; Vandesompele et al. 2002). OsGADPH was identified as most stable gene when assessed through NormFinder and ΔCT, while geNorm identified Osβ-TUB and OseIF4α as the most stable. Variation in the ranking of the gene, as evident from our study, could be attributed to the use of different algorithm programs such as geNorm and NormFinder (Vandesompele et al. 2002; Andersen et al. 2004). Such discrepancies were also reported while identifying OseIF4α as the most stable housekeeping gene in pigeon pea under drought stress using similar statistical method (Sinha et al. 2015). NormFinder and GeNorm algorithms have been previously reported in soybean for identification of Elongation factor 1-beta and ACT11 as stable internal reference gene during anoxic stress (Nakayama et al. 2014).
Variation in stability of known housekeeping genes under the hypoxic condition is not unexpected as similar findings have been reported for genes used as internal reference control, like GAPDH, β-Actin 11, β-TUBLINE, PGK,UBQ, RPL-19, and 18S rRNA in a wide variety of tissues and cells (Rebouças et al. 2013). Importantly, our analysis suggests that housekeeping genes like Os25SrRNA should be avoided as an internal control for studying the expression profile of candidate genes under hypoxic conditions in rice. This is because the expression of endogenous target gene OsTPP7 normalized with Os25SrRNA was lower in rice genotypes tested under different hypoxic treatments. Suboptimal performance of Os18SrRNA and Os25SrRNA gene had been reported previously while using them as internal controls for normalizing the drought-induced gene uspA (Sinha et al. 2015). Finally, comparative study on expression dataset obtained from four different statistical methods using RefFinder (Mallona et al. 2010) and ranking the genes based on geo mean also identified OsGADPH as the most stable internal control gene under hypoxia.
It has been previously reported that the performance and gene stability of certain housekeeping genes may get altered depending on the type of treatment. The stable housekeeping gene (OsGAPDH) identified through normalization of candidate genes in the present study is known to be cytosolic glyceraldehyde 3-phosphate dehydrogenase (OsGAPDH, E.C. 1.2.1.12), present in all organisms and catalyzes the oxidation of trios phosphate during glycolysis (Giulietti et al. 2001). Some commonly used reference genes like ACT 11 and OsGAPDH performed inconsistently in different plants, tissues, and experimental conditions, and that warrants for caution in its use as an internal control, primarily because transcript levels of reference genes could vary considerably in response to experimental conditions, cellular process, and tissue types. It is also true that OsGAPDH acts as a component of the glycolytic pathway, and gluconeogenesis, processes that are integral to the survival of the organism. Yet, studies in other crop species have shown that OsGAPDH has constant expression in total developmental stages in crops like pearl millet (Reddy et al. 2015) and sugarcane (Larissa Mara de Andrade et al. 2016) while being the least stable transcript in wheat developmental series. Also OsGAPDH was reported as an appropriate candidate for reliable normalization of target gene expressions levels during biotic and abiotic stresses in rice (Bevitori et al. 2014). Although it is reported that expression level of 37 kDa OsGAPDH subunit changes during early stages of germination during anoxia (Ricard et al. 1989), in our study, we focused on hypoxic condition (2–3% oxygen level) for reference genes whose transcript does not vary in tolerant and susceptible rice cultivars under hypoxia germination stage oxygen deficiency (GSOD). Although OselF-4a and OselF-1a were found to be stable besides OsGAPDH in GSOD-tolerant and susceptible cultivar in hypoxia, cytosolic form OsGAPDH2 was ranked as most stable when all the datasets from different algorithms were analyzed together. The transcript level of target gene OsTPP7 was also expectedly found higher in GSOD-tolerant rice germplasm while using OsGAPDH as an internal control. For our study, we used OsGAPDH2 cytosolic form located on the chromosome 4, (Locus: chr04:24305454.24309055) as there is precedence for its use in reference gene evaluation in rice (Jain et al. 2006). The OsTPP7 has been used as a hypoxic germination biomarker gene in the present study. High fold change in expression of OsTPP7 gene, normalized against stable endogenous candidates compared to less stable ones, across different time intervals in tolerant cultivar was also in tune with the functional aspect of the gene. It would be of interest to investigate in future if the selected endogenous genes could maintain similar stability and performance levels when validated with other biomarkers of hypoxic germination like Ramy3D. Notwithstanding the effect of stable candidate endogenous control on other hypoxic biomarker genes, it would be safe to assume that the most stable housekeeping genes identified from the combination of the algorithms can be used as an internal control for global transcript study during hypoxic germination of rice.
Conclusion
We have compared and ranked the performance of reference genes in RT-qPCR using three different germplasms (2 hypoxic tolerant and 1 susceptible) under hypoxic condition, analyzed using RefFinder. Reliability of two genes, namely OsGAPDH and OsEF1α, for its use as endogenous control was further validated through expression of known hypoxia-induced gene OsTPP7 during germination in tolerant germplasms. Our findings suggest that the identified reference genes OsGAPDH and OsEF1α could be considered as the most stable housekeeping genes for expression studies in rice germination studies under hypoxia.
References
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Aniko S, Charles M, Mehmet K et al (2004) Statistical modeling for selecting housekeeper genes. Genome Biol 5:R59
Bevitori R, Oliveira MB, Grossi-de-Sá MF et al (2014) Selection of optimized candidate reference genes for qRT-PCR normalization in rice (OryzasativaL.) during Magnaportheoryzaeinfection and drought. Genet Mol Res 13(4):9795–9805
Bo J, Bin L, Yurong B et al (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9:59
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
Chi X, Hu R, Yang Q et al (2012) Validation of reference genes for gene expression studies in peanut by quantitative real-time RT-PCR. Mol Gen Genomics 287:167–176
Christianson JA, Llewellyn DJ, Dennis ES Wilson IW (2010) Global gene expression responses to waterlogging in roots and leaves of cotton (GossypiumhirsutumL.). Plant Cell Physiol 51:21–37
Dumbala S, Pooja B, Katamreddy C, Kiran S (2013) Evaluation and validation of reference genes for normalization of quantitative real-time PCR based gene expression studies in peanut. PLoS One 8(10):e78555
Giulietti A, Overbergh L, Valck D et al (2001) An overview of real time quantitative PCR: applications to quantify cytokine gene expression. Methods 25:386–401
Hu R, Fan C, Li H et al (2009) Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Bio 10:93
Isaiah M, Naoki Y, Jennylyn L, Toshisangba L (2016) Validation of reference genes for gene expression analysis in switchgrass (Panicum virgatum) using quantitative real-time RT-PCR. PLoS One 9(3):e91474
Jackson MB, Ram PC (2003) Physiological and molecular basis of susceptibility and tolerance of rice plants to complete submergence. Ann Bot 91:227–241
Jain M, Nijhawan A, Tyagi AK et al (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651
Kretzschmar T, Pelayo MAF, Trijatmiko KR et al (2015) A trehalose-6-phosphate phosphatase enhances hypoxic germination tolerance in rice. Nat Plants 1:15124
Kreuzwieser J, Hauberg J, Howell KA et al (2009) Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiol 149:461–473
Lasanthi-Kudahettige R, Magneschi L, Loreti E et al (2007) Transcript profiling of the anoxic rice coleoptile. Plant Physiol 144:218–231
Le DT, Aldrich DL, Valliyodan B et al (2012) Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PLoS One 7:e46487
Liu F, Vantoai T, Moy LP et al (2005) Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol 137:1115–1129
Long XY, Wang JR, Ouellet T et al (2010) Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant MolBiol 74:307–311
Ma S, Niu H, Liu C et al (2013) Expression stabilities of candidate reference genes for RT-qPCR under different stress conditions in soybean. PLoS One 8:e75271
Maksup S, Supaibulwatana K, Selvaraj G (2013) High-quality reference genes for quantifying the transcriptional responses of Oryzasativa L. (ssp. indica and japonica) to abiotic stress conditions. Chin Sci Bull 58:1919–1930
Mallona I, Lischewski S, Weiss J et al (2010) Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol 10:4
Manoli A, Sturaro A, Trevisan S et al (2012) Evaluation of candidate reference genes for qPCR in maize. J Plant Physiol 169:807–815
Ming-Le W, Qing-Hui L, Hua-Hong X et al (2017) Reliable reference genes for normalization of gene expression data in tea plants (Camelliasinensis) exposed to metal stresses. PLoS One 12(4):e0175863
Mustroph A, Lee SC, Oosumi T et al (2010) Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol 152:1484–1500
Nakayama TJ, Rodrigues FA, Neumaier N et al (2014) Reference genes for quantitative real-time polymerase chain reaction studies in soybean plants under hypoxic conditions. Genet Mol Res 13(1):860–871
Nolan T, Hands RE, Bustin SA (2006) Quantification of mRNA using real-time RT-PCR. Nat Protocol 1:1559–1582
Ohl F, Jung M, Xu C (2005) Gene expression studies in prostate cancer tissue: which reference gene should be selected for normalization. J Mol Med 83:1014–1024
Paolacci AR, Tanzarella OA, Porceddu E et al (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol 10:11
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515
Rebouças EL, Costa JJN, Passos MJ et al (2013) Real time PCR and importance of housekeepings genes for normalization and quantification of mRNA expression in different tissues. Braz Arch Biol Technol 56(1)
Reddy DS, Bhatnagar-Mathur P, Cindhuri KS et al (2013) Evaluation and validation of reference genes for normalization of quantitative real-time PCR based gene expression studies in peanut. PLoS One 8:e78555
Reddy PS, Reddy DS, Sharma KK et al (2015) Cloning and validation of reference genes for normalization of gene expression studies in pearl millet [Pennisetumglaucum (L.) R. Br.] by quantitative real-time PCR. Plant Gene:35–42
Reena N, Katharine AH, Adam C et al (2009) Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol 151:306–322
Reena N, David S, Matthew D et al (2017) Dynamic and rapid changes in the transcriptome and epigenome during germination and in developing rice (Oryzasativa) coleoptiles under anoxia and re-oxygenation. Plant J 89:805–824
Ricard B, Rivoal J, Pradet A (1989) Rice cytosolic glyceraldehyde 3-phosphate dehydrogenase contains two subunits differentially regulated by anaerobiosis. Plant Mol Biol 12(2):131–139
Sasidharan R, Mustroph (2011) A plant oxygen sensing is mediated by the N-end rule pathway: a milestone in plant anaerobiosis. Plant Cell 23(12):4173–4183
Selvey S, Thompson EW, Matthaei K et al (2005) Beta-Actin 11—an unsuitable internal control for RT-PCR. Mol Cell Probes 15(5):307–311
Septiningsih EM, Carlos IJ, Ignacio et al (2013) QTL mapping and confirmation for tolerance of hypoxic conditions during germination derived from the rice landrace Ma-Zhan red. Theor Appl Genet 126:1357–1366
Setter TL, Ellis M, Laureles CV, Ella ES et al (1997) Physiology and genetics of submergence tolerance in rice. Annu Rev Bot 79:67–77
Silver N, Best S, Jiang J, Thein SL (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7:33
Sinha P, Singh VK, Suryanarayana V et al (2015) Evaluation and validation of housekeeping genes as reference for gene expression studies in pigeonpea (Cajanuscajan) under drought stress conditions. PLoS One 10(4):e0122847
Thellin O, Zorzi W, Lakaye B et al (1999) Housekeeping genes as internal standards: use and limits. J Biotechnol 75:291–295
Vandesompele J, De PK, Pattyn F et al (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):RESEARCH0034
Ward D, Jutta D, Roswitha W et al (2015) Reference gene validation for RT-qPCR, a note on different available software packages. PLoS One 10(3):e0122515
Xie F, Sun G, Stiller JW, Zhang B (2011) Genome-wide functional analysis of the cotton transcriptome by creating an integrated EST database. PLoS One 6(11):e26980
Xu M, Zhang B, Su X et al (2011) Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal Biochem 408:337–339
Xu H, Bau J-D, Dai J-S (2015) Genome-wide identification of new reference genes for qRT-PCR normalization under high temperature stress in rice endosperm. PLoS One 10(11):e0142015
Yueai L, Chenlu Z, Hai L, Shibin G et al (2014) Validation of potential reference genes for qPCR in maize across abiotic atresses, hormone treatments, and tissue types. PLoS One 9(5):e95445
Zou X, Jiang Y, Liu L et al (2010) Identification of transcriptome induced in roots of maize seedlings at the late stage of waterlogging. BMC Plant Biol 10:189
Funding
This work was financially supported by the Department of Biotechnology, Government of India.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: DK, PKD, and BKS. Performed the experiments: DK. Analyzed the data: DK, PKD, and BKS. Wrote the manuscript: DK, PKD, and BKS. Corrected the manuscript: PKD and BKS.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by: Barbara Naganowska
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kumar, D., Das, P.K. & Sarmah, B.K. Reference gene validation for normalization of RT-qPCR assay associated with germination and survival of rice under hypoxic condition. J Appl Genetics 59, 419–430 (2018). https://doi.org/10.1007/s13353-018-0466-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-018-0466-1