Abstract
Rice oxalate oxidase genes (OXO) may play a role in resistance to Magnaporthe oryzae. Genome analyses showed four tandemly duplicated OXO genes, OsOXO1–OsOXO4, which mapped to a blast resistance QTL in chromosome 3. These genes have >90% nucleotide and amino acid identity, but they have unique gene structures, conserved motifs, and phylogeny compared to the 70 other members of the cupin superfamily in the Nipponbare genome, which were divided into several classes. In resistant and susceptible Vandana/Moroberekan advanced backcross lines, only OsOXO4 was expressed during rice–M. oryzae interactions, and its expression increased earlier in resistant than susceptible lines. The earlier expression of OsOXO4 in resistant lines correlated with a 26-bp promoter insertion containing an additional copy of the bacterial responsive nodulation cis-element. Our results showed that OsOXO1–4 are in a separate class of rice cupin genes and supports a role for the promoter variant of OsOXO4 in resistance to M. oryzae.
Similar content being viewed by others
Introduction
One of the largest groups of proteins are cupins, which are a superfamily of functionally diverse but structurally related proteins (Dunwell et al. 2001; Dunwell et al. 2008). Two conserved motifs, ([G(X)5HXH-(X)3,4E(X)6G] and G(X)5PXG(X)2H(X)3N]), are shared by all cupins, and together make up the cupin domain. Two His residues and a Glu residue in Motif 1, together with a His residue in Motif 2, act as ligands for the binding of a manganese ion at the active site of the archetypal cupin, germin (Gane et al. 1998). Between the two cupin motifs is an intermotif region, that varies in length from 15 residues in many bacterial cupins to more than 50 residues in some plant cupins, and the number of residues in the intermotif region have been used to define at least 18 subclasses of cupins from archaea, bacteria, and eukaryotes (Dunwell et al. 2000). In addition to subclasses, cupins can be divided into two groups, the monocupins and bicupins (Dunwell et al. 2004). Monocupins, such as bacterial phosphomanose isomerases (PMIs), bacterial AraC transcriptional regulators, plant oxalate oxidases (OXO), and plant germins, have a single copy of the cupin domain, while bicupins, such as the plant seed storage proteins and plant oxalate decarboxylases, have two copies (Khuri et al. 2001).
Germin was first discovered in wheat during the onset of germination and was later determined to have similarity with barley OXOs that are responsible for the degradation of oxalate (Lane et al. 1993). Cereal germins and OXOs cluster together in phylogenetic analyses and are considered to be the true germins (Carter and Thornburg 2000; Khuri et al. 2001). OXOs may be important in disease resistance because they degrade oxalate, and oxalate is a virulence factor produced by several fungal pathogens (Bateman and Beer 1965; Godoy et al. 1990; Cessna et al. 2000; Guimaraes and Stotz 2004; van Kan 2006). In addition to degrading oxalate, OXO activity generates H2O2 from the reaction of oxalate + O2 + 2 H+ = 2 CO2 + H2O2 (Lane 1994). Active oxygen species, such as H2O2, are reported to be involved in plant defense responses (Lamb and Dixon 1997). This can be through oxidative cross-linking of lignin and proteins during cell wall modification, activating gene expression for transcription-dependent defenses, and acting directly as antimicrobial agents to inhibit pathogen development (Lamb and Dixon 1997). OXO-generated H2O2 may also function as a secondary messenger in the activation of phytoalexin biosynthetic pathways, hypersensitive response (HR), systemic acquired resistance, and PR gene expression in plants, all of which have been related to disease resistance (Carter et al. 1998).
Genes with sequence similarity to germins are termed germin-like proteins (GLPs), which have sequence similarity to OXO but are not true germins (Membre et al. 1997; Membre et al. 2000; Khuri et al. 2001). GLPs have a variety of functions (Zhou et al. 1998; Faris et al. 1999; Druka et al. 2002; Ramalingam et al. 2003; Liu et al. 2004; Wu et al. 2004; Manosalva et al. 2008). For example, a GLP, PSGER1, in peas shows sequence similarity to the N-terminal sequence of a putative plant receptor for rhicadhesin, a bacterial attachment protein (Gucciardo et al. 2007). Such rhicadhesin receptor-like GLPs have been found to be strongly induced in roots and root cultures of Medicago truncatula colonized by the arbuscular-mycorrhiza (AM) fungus, Glomus intraradices (Doll et al. 2003). Another example of a GLP is the Nectarin I precursor protein, which is located in floral nectaries of tobacco where it exhibits superoxide dismutase activity that generates high levels of hydrogen peroxide in the nectar and thus may function in plant defense (Carter and Thornburg 2003). Other GLPs have also been implicated in a variety of plant–microbe interactions. For example, an OXLP was associated with papilla formation in barley during infection with Blumeria (Erysiphe) graminis f.sp. hordei (Bgh) (Wei et al. 1998).
OXOs and GLPs have been implicated in quantitatively inherited disease resistance in rice (Faris et al. 1999; Ramalingam et al. 2003; Liu et al. 2004; Wu et al. 2004). In some rice germplasms, GLPs contributed to a major effect QTL on chromosome 8. Gene silencing studies recently demonstrated that a cluster of chromosome 8 GLPs contributed to broad-spectrum, basal disease defense responses (Manosalva et al. 2008). In other germplasms, OXOs were the major contributors to rice blast resistance (Ramalingam et al. 2003; Wu et al. 2004). OXO was mapped to a QTL in chromosome 3 of rice and was also shown to contribute to variation in number of rice blast lesions in an advanced backcross population derived from rice cultivars Vandana and Moroberekan (Wu et al. 2004). Moroberekan is a tropical japonica variety from Africa that is highly resistant to rice blast (Wang et al. 1994) while Vandana is a rice blast-susceptible cultivar from India (Wu et al. 2004). Thus, OXO genes have been associated with blast resistance.
Although members of the cupin gene family are important in a variety of plant functions, including disease resistance, only a few specific gene family members had previously been studied in rice and no systematic phylogenetic analysis of the family was available. The publication of a high quality, complete genome sequence for rice (IRGSP 2005) enabled a characterization of the entire cupin gene family of rice. In the process, we identified four OXO genes (OsOXO1, OsOXO2, OsOXO3, and OsOXO4) on chromosome 3 of the Oryza sativa ‘Nipponbare’ genome that co-localized with a previously reported blast disease resistance QTL (Ramalingam et al. 2003; Wu et al. 2004). All four OXOs exhibited similar structural gene sequences, but the promoter regions were distinct from each other. To determine if promoter differences could account for phenotypic differences in blast disease resistance, we compared promoters of the four genes from both Moroberekan and Vandana and progenies from an intermated population. We demonstrated that only the promoters of OsOXO4 from resistant varieties contained an extra element suggesting differential regulation of the genes. Consistent with this prediction, we show a correlation with OsOXO4 expression and disease resistance.
Results
Phylogenomic analysis of rice cupins
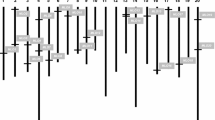
Seventy cupin sequences were retrieved from the genome sequence of O. sativa cv. Nipponbare using a PERL script designed to identify conserved protein sequences with the cupin domains, [G(X)5HXH(X)3,4E(X)6G] and G(X)5PX(G)2H(X)3N]. Phylogenetic analysis divided the cupin sequences into clusters corresponding to the monocupins and bicupins (Fig. 1), and this division was confirmed using the CD-Search option of GenBank’s Conserved Domain Database (Marchler-Bauer et al. 2005). There were 25 bicupins that were divided into three clusters corresponding to the annotation designations at SWISS-PROT: glutelin precursor proteins, globulin-1 proteins, and 12S seed storage proteins (as shown in Supplementary Table 1). The glutelin precursor proteins were more closely related to the globulin-1 proteins than the 12S seed storage proteins, which comprised the largest group of bicupin genes (Fig. 1). There were 45 monocupins divided into two large clusters (Fig. 1). One cluster contained all GLPs similar to barley OXLPs, which was termed the OXLP cluster, and the other was the more diverse non-OXLP cluster. The non-OXLP cluster was divided into five sub-clusters, termed subclasses, which corresponded to their SWISS-PROT annotations (Fig. 1; as shown in Supplementary Table 2). These five subclasses were tentatively defined as OXOs, rhicadhesin receptor-like GLPs, ‘unknown function’ GLPs, nectarin 1 precursor-like proteins and auxin-binding-like proteins (ABPLs). The clustering of these protein sequences indicates that they are more closely related to each other than to the OXLPs. In addition to the two cupin domain motifs, a common feature of these genes is the (L/V)QDFCV(A/G) motif, which is strongly conserved among only monocupins (Membre et al. 1997).
Phylogenetic relationships of protein sequences with the cupin domain PFAM00190 in the genome of rice cultivar Nipponbare. The tree was generated by distance matrix using ClustalX and rooted with spherulin a. The bicupin class contains 25 genes which were all annotated as seed storage proteins. The monocupin class is composed of 45 genes; the annotation of these genes is shown at the right of the figure. Homologous genes physically close to each other are joined by bars or bars joined by brackets. Genes with expression evidence are boldfaced. Genes marked with an asterisk do not belong to any subclass or cluster and are considered as phylogenetic singletons. The values to the left of the subclass are the results of positive selection analyses, where p is the probability value, ω is the dN/dS value under M8 of codeml, and P1 is the inferred proportion of positively selected sites.
While a number of monocupin genes occurred as singletons in the O. sativa cv. Nipponbare genome, most members were in eight clusters of two or more genes on chromosomes 1, 3, 4, 5, 8, 9, and 12. Clustering on chromosomes correlated with the monocupin subclass (Fig. 1) and often occurred in tandem arrays. All four OXO genes in rice are tandemly arrayed on chromosome 3 and have >90% AA identity with most of the variation at the 5’ end (Supplementary Fig. 1). Similarly, the six rhicadhesin receptor-like GLP genes occurred as a tandem pair on chromosome 1, two singletons on chromosome 1, and a singleton each on chromosome 3 and 5. The nectarin 1 precursor-like protein genes occurred as three tandem genes on chromosome 3 and a singleton in chromosome 1, the ABPLs occurred as tandem pair on chromosome 8 and a singleton on chromosome 4, and the ‘unknown function’ GLPs were a tandem pair on chromosome 9 with three singletons on chromosomes 2, 3, and 5. The OXLPs were three tandem genes on chromosomes 12 and 2, and 11 tandem genes on chromosome 8, as well as two singleton OXLPs on chromosomes 4 and 8. The singleton (LOC_Os08g13440) in chromosome 8 is 2.7 Mb downstream of the cluster of 11 tandem OXLP genes and groups phylogenetically with two OXLPs in the cluster (Fig. 1).
The high occurrence of tandem arrays indicates gene duplication, which is supported by all the genes being oriented in the same direction in the eight clusters with two or more monocupin genes. One exception may be the singleton OXLP (LOC_Os08g13440) on chromosome 8. Although LOC_Os08g13440 has ∼80% AA identity with the OXLPs in the tandem array, it is oriented in the opposite direction (as shown in Supplementary Table 2).
Protein alignment and analyses of the rice monocupins
Alignment of the 45 monocupin AA sequences from rice showed a total of 25 motifs identified by MEME analysis (Fig. 2; as shown in Supplementary Table 3). All subfamily members contain motifs 1 to 8, which form the conserved cupin domain (PF00190) (Fig. 2; as shown in Supplementary Table 3), and motifs 1 and 2 contain the active sites of the cupin domain (Khuri et al. 2001; Dunwell et al. 2004). The sequence of the intermotif region between Motif 1 and 2 is consistent within subclasses identified by phylogenetic analyses, except for the rhicadhesin receptor precursor subfamily, in which LOC_Os01g72300 has 22 AA instead of the 18 AA characteristic for this subclass (as shown in Supplementary Fig. 2).
Hydrophobicity and protein motif in the monocupins. Sequence of proteins is based on the monocupin gene tree. Hydrophobicity was calculated using the Kyte-Doolittle algorithm. Protein motifs were identified using MEME (http://meme.sdsc.edu/meme/intro.html).
While motifs 1–8 defined the cupin domain, motifs 9–25 did not define another cluster of orthologous gene (COG) domain. Rather, these motifs corresponded to the subclasses identified in Fig. 1 that corresponded to the SWISS-PROT annotations. Certain motifs or combinations of motifs were unique to subclasses of monocupins (Fig 2; as shown in Supplementary Table 3). Motif 9 is only found in nectarin I precursor-like proteins and OXLPs. Motif 11 is only found in OXLPs in chromosomes 8 and 12 but not in the three OXLPs on chromosome 2. Motifs 13, 14, and 16 are unique among OXOs, and motif 15 is unique to the ABP-like proteins and rhicadhesin receptor-like proteins. Motifs 17 and 19 are found only in the GLP subclass with unknown functions.
Some motifs are unique to cereals, i.e., motifs 9–12 and 17–19 were identified only in cereal GLPs and motifs 13–14 and 16 were found only in cereal OXOs. On the other hand, motifs 15 and 20 were annotated as ABP/adenosine diphosphate glucose pyrophosphatase (ADP) (as shown in Supplementary Table 3), which is common to germins in all flowering plants (Rodriguez-Lopez et al. 2001). Other motifs were not unique to cupins, i.e., motifs 21 and 23–25 matched hypothetical proteins, while motif 22 is similar to a retrotransposon gag protein (Marchler-Bauer et al. 2005).
All monocupins contain a signal peptide, except LOC_Os03g58980 and LOC_Os03g58990, which are annotated as nectarin 1 precursor protein GLPs (as shown in Supplementary Table 4). The OXOs and GLPs have distinct signal peptides (motif 14 in the OXOs and motif 10 in the OXLPs; Fig. 2). Signal peptides predicted by SignalP for the monocupins in the other subclasses were not well enough conserved to be identified as motifs by MEME analysis. All monocupins were predicted by Wolf pSORT to be located in the extracellular matrix. The only monocupins with the same signal peptide sequence were OsOXO3 and OsOXO4 (as shown in Supplementary Fig. 1).
Hydrophobicity profiles of the monocupins correspond with the motifs identified by MEME analysis (Fig. 2). A hydrophobic peak corresponding to the signal peptide sequence is followed by a relatively neutral region and then a series of three or more hydrophobic peaks corresponding to motifs 3, 1, 7, and 2. Hydropathy profiles were similar within subclass members, except for LOC_Os02g29010 and LOC_Os04g22080, which were in the OXLP subclass and have transposon insertions (Fig. 2). LOC_Os03g58990 in the nectarin I precursor-like protein subclass (Fig. 1) lacks several motifs and also has a hydrophobicity profile distinct from the rest of the monocupins. The predicted isoelectric points for the monocupin proteins range from 5.12–9.48, but differences in pI did not correspond to any of the monocupin subclasses (Fig. 2). A 1–2 N-glycosylation is a characteristic of all monocupins (Membre et al. 2000) and was found for all the rice monocupins, except LOC_Os04g52720 and LOC_Os8g35750, which have 1–4 N-glycosylation sites (as shown in Supplementary Table 4).
Gene structure of monocupins
Of the 45 monocupin genes, 15 have no intron, 27 have one intron, and three have multiple introns (as shown in Supplementary Fig. 3). Interestingly, none of the four OXOs have introns, and the rhicadhesin receptor-like proteins all have one intron. The presence of untranslated regions (UTRs) varied among the monocupin genes (as shown in Supplementary Table 5). However, a search using KOME (http://cdna01.dna.affrc.go.jp/cDNA/) showed that full-length cDNAs are only available for the 19 monocupin genes with both 5’ and 3’ predicted UTRs. A search of the GenBank EST database dbEST (http://www.ncbi.nlm.nih.gov/dbEST/index.html) containing over 28,262,948 ESTs showed that genes with both 5’ and 3’ predicted UTRs have many more ESTs (average = 65 ESTs) than genes with either a 5’ or 3’ UTR only (average = 6 ESTs) or no UTRs at all (average = 3 ESTs) (as shown in Supplementary Table 5).
Selection pressure among monocupins
From the ratio of synonymous to nonsynonymous substitution, selection pressure could not be detected among monocupin genes in the different subclasses, except for the rhicadhesin-like receptor protein, OXOs and OXLPs (Fig. 1). Diversifying selection was detected in rhicadhesin-like receptor protein subclass (ω = 1.1257), while the OXOs are under purifying selection (ω = 0.1361) (Yang et al. 1998). The OXLPs are divided into three groups based on phylogenetic clustering (Fig. 1), but all three groups are under purifying selection (ω = 0.1397 0.2072 and 0.1273).
OXOs are unique in cereals
BLAST search against the GenBank non-redundant database using the rice OXOs showed that similar genes (e> 8e-132) are found in cereals only. The OXOs belong to family ID 1869 (http://www.phytome.org) which is divided into two subfamilies (Fig. 3). Subfamily 1 has two clusters, one with only the four rice OXOs and the other cluster composed of OXOs from maize, wheat, switch grass, and sorghum. OsOXO2, OsOXO3, and OsOXO4 are 74% similar with HvOXOa while OsOXO1 shares 72% similarity with HvOXOa. Subfamily 2 is composed of oxalate oxidase genes from wheat and barley.
Phylogenetic relationships of cereal oxalate oxidases. The cereal oxalate oxidases are classified into two subfamilies based on amino acid sequences. Subfamily 1 consists of rice, wheat, switchgrass, sorghum, and maize sequences. All rice oxalate oxidases belong to Subfamily 1 and sequences osat203469, osat169958, osat176731, and osat168006 correspond to LOC_03g48780, LOC_03g48770, LOC_03g48760 and LOC_03g48750, respectively. Subfamily 2 consists of wheat and barley oxalate oxidases. Amino acid sequences were derived from Phytome (http://www.phytome.org).
Comparison of OXO sequences in resistant and susceptible rice-blast interactions
Rice cultivar Moroberekan is a donor of disease resistance QTL in crosses with Vandana (susceptible) and one of the disease resistance QTL was demonstrated to co-localize with the chromosome 3 OXO genes (Wu et al. 2004). To determine if differences among the sequences of the four OXO genes on chromosome 3 might correlate with phenotypic differences in disease responses, we sequenced the four OXO genes from cvs. Moroberekan and Vandana. In addition, we sequenced alleles of the genes from progeny lines of a cross between Vandana and Moroborekan (Table 1). Resistant lines IR78221-19-6-7-B, IR78221-19-6-56-B, and IR78221-19-6-99-B were demonstrated by molecular marker analysis to have the OXO QTL fragment from Moroberekan (Carrillo and Vera Cruz, unpublished data). In contrast, line IR78221-19-3-196-B has the OXO QTL fragment from Vandana and is susceptible to rice blast under field conditions in India and the Philippines (Table 1).
We observed several nucleotide differences in the OXO genes between cvs. Moroberekan and Vandana that resulted in AA differences. In OsOXO1, there was a difference at AA 123 with an L in Moroberekan and a V in Vandana (as shown in Supplementary Fig. 4). In OsOXO2, a CG insertion at nucleotide position 122 in Vandana results in a stop codon for Vandana, but not for Moroberekan, making its protein 174 AA shorter (as shown in Supplementary Fig. 5). OsOXO3 was identical in Moroberekan and Vandana. In OsOXO4, there was a difference of F to Y at AA 37, V to D at AA 46, and V to I at AA 117 between Moroberekan and Vandana, respectively (as shown in Supplementary Fig. 4). The difference at AA117 is in motif 1 of the cupin domain. For all four OXO genes, the resistant progenies, IR78221-19-6-7-B, IR7822-19-6-56-B, and IR78221-19-6-99-B, have the same sequence as Moroberekan, while the susceptible progeny, IR78221-19-3-196-B, has the same sequence as Vandana.
While the structural genes were quite similar among the parents and six progeny, comparison of the 1,000-bp upstream regions of OsOXO4 from the six lines showed that they had 96% nucleotide identity, except for a 26-bp region in Moroberekan and the resistant progenies that was absent in Vandana and the susceptible progeny (Fig. 4). Cis-elements identified within this 26-bp sequence included four copies of bacterial responsive nodulation genes, one copy of a cis-element related to the DNA-binding with one finger (DOF) protein binding site in maize, and a copy of a cis-element similar to over-represented sequences in light-induced promoters in Arabidopsis (Wojtaszek 1997).
Pairwise alignment of the 1,000-bp upstream region of OsOxO4 from Moroberekan and Vandana showing the 26-bp difference between the two sequences in the region from −353 to −378 in the Moroberekan sequence. Below the alignment are the locations of the cis-elements in this region. These are the bacterial-responsive nodulation elements, NODCON2GM and OSE2ROOTNODULE, the binding site for the DOF proteins in maize, DOFCOREZM, and the light-induced promoter element SORLIP1AT. The 26-bp sequence is present in Moroberekan and the blast resistant progenies IR78221-19-6-7-B, IR78221-19-6-56-B and IR78221-19-6-99-B, but is absent from Vandana and the blast susceptible progeny IR78221-19-3-196-B. Minus symbol indicates that the promoter element is on the opposite strand.
Expression of the OsOXO4 is correlated with quantitative disease resistance to blast
We tested the expression of the chromosome 3 OXO genes in Vandana and Moroborekan as well as in their progeny to determine if expression of any member correlated with involvement in quantitative resistance. The detection of the promoter element we described above in OsOXO4 in Moroborekan (harbors chromosome 3 QTL for blast resistance) but not in Vandana (no chromosome 3 QTL) cultivars and their progeny lines suggested that differential regulation of the gene might correlate with host responses involved in quantitative resistance. Indeed, using reverse transcription polymerase chain reaction (RT-PCR), we detected OsOXO4 transcripts, but not OsOXO1, OsOXO2, and OsOXO3 after inoculation with M. oryzae. OsOXO4 was expressed in Moroberekan, IR78221-19-6-7-B, IR78221-19-6-56-B, and IR78221-19-6-99-B at 24 h after inoculation with M. oryzae PO6-6 but was not detected until 48 h after inoculation in the susceptible interactions with Vandana and IR78221-19-3-196-B (Fig. 5).
RT-PCR analysis of the expression profiles of Vandana, IR78221-19-3-196-B, IR78221-19-6-7-B, IR78221-19-6-56-B, and IR78221-19-6-99-B and Moroberekan using gene specific primers for four OXO genes. Each line was inoculated with Magnaporthe oryzae strain PO6-6 (100,000 spores/ml). Leaf samples were collected at 0-, 6-, 12-, 24-, and 48-h post-infection (HPI) and extracted for mRNA. A ∼300-bp transcript was amplified from OsOxO4.
Fifteen EST libraries from the DFCI Rice Gene Index (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=Rice) were surveyed for the expression of each OXO gene by BLAST. Each gene had different temporal and spatial expression patterns except for OsOXO2 where no EST was detected under any condition (as shown in Supplementary Fig. 6). OsOXO1 was expressed in the panicles and at the flowering and pollination stages. OsOXO3 ESTs were found in roots and seed libraries. OsOXO4 ESTs were detected in drought, cold stress, and metal (CuSO4) stress libraries, as well as in healthy root, shoot, and leaf libraries. It was also expressed at tillering and booting stages (as shown in Supplementary Fig. 6). These results are consistent with a role for OsOXO4 in rice stress in general.
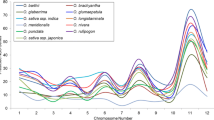
We also surveyed the Rice massively parallel signature sequencing (MPSS) database for expression of the four OsOXOs (Meyers et al. 2004a). MPSS signatures were only detected for the OsOXO4 in 23 of 70 libraries (Fig. 6). The MPSS signature for OsOXO4 was also correlated with the available EST data. The OsOXO4 signature was detected in libraries for germinating seedlings, mature leaves as well as in young leaves stressed with 250 mM NaCl for 24 h (data not shown). Of relevance to this study, OsOXO4 was induced by inoculation with both M. oryzae and Xanthomonas oryzae pv. oryzae (Xoo) and after insect or mechanical damage.
MPSS expression profile of OsOxO4 plotted in parts per million of transcripts in rice-Magnaporthe oryzae (Mo) susceptible (Mo-S) and resistant (Mo-R) interactions and in rice-Xanthomonas oryzae pv. oryzae (Xoo) susceptible (Xoo-S) and resistant interaction (Xoo-R). Leaf samples for rice-Mo interaction were collected Nipponbare (susceptible) and Nipponbare-Pi9 (resistant) inoculated with Mo. Leaf samples for rice-Xoo interaction were collected from Nipponbare (susceptible) and Nipponbare-Xa21 (resistant) inoculated with Xoo.
Discussion
Most of the known genes involved in QTL-based defense responses are members of complex gene families. This poses difficulties in utilizing them in practical plant breeding. The approach described in this work to elucidate the functional OXO in rice resistance to M. oryzae combined genomic information with phenotypic selection, allowing us to narrow down the target chromosomal region for disease resistance and to identify features that distinguish that gene from other members of the cupin superfamily in rice. A phylogenetic analyses of the OXOs in relation to the cupins in rice showed that it is a sizable gene family with 70 members. The family could be divided into two major groups, the monocupins and the bicupins, and with 45 members, the monocupins are the larger group. The monocupins are comprised of germins and GLPs while the bicupins are predicted to be seed storage proteins (Dunwell et al. 2004). Our study focused on the OXO genes that belong to a subclass of the monocupins and are predicted to be involved in disease defense responses.
Excluding the OXLP cluster, which were more distinct, there are five monocupin subclasses (>50% nucleotide and AA identity). Most of the members of the subclasses occur in tandem clusters in the same genomic loci, suggesting that these arose from tandem gene duplications, where a sequence of DNA is transferred to create two adjacent copies (Elemento et al. 2002). This is consistent with observations for Arabidopsis, where the level of tandem gene duplication in the germin family is more than two standard deviations above the genome median (Cannon et al. 2004). Our analyses show that the OXO mapped to a QTL in chromosome 3 associated with resistance to rice blast (Ramalingam et al. 2003; Wu et al. 2004) contains four tandemly duplicated OXOs in a 10 kb region. While the gene-coding region of the four OXO genes have >90% nucleotide identity, analyses of their 1,000-bp upstream regions within Nipponbare show little nucleotide identity, which may affect how duplicate genes function in nature. Gene duplication has been predicted to create gene diversity because the duplicated genes can diverge to acquire new functions (Ohno 1970; Roth et al. 2006). However, most eukaryotic genome studies show that the sequence of the duplicated genes do not diverge as much as expected (Force et al. 1999; Lynch et al. 2001). The duplication–degeneration–complementation model of gene duplication (Force et al. 1999) predicts that degenerative mutations in regulatory elements can increase (rather than reduce) the probability of duplicate gene preservation and predicts that the usual mechanism of duplicate gene preservation is the partitioning of ancestral functions rather than the evolution of new functions. This is true for the rice monocupin family, where the duplicated genes retain relatively high nucleotide identity.
A survey of the expression of monocupins in EST libraries (http://www.tigr.org/tdb/e2k1/osa1/locus_expression_evidence.shtml) indicates a very wide range of expression patterns and indicates that many gene family members are functional. By correlating available EST data with gene structure, only genes predicted to have both 5’ and 3’ UTRs are expressed, although some members with only one UTR have ESTs available. For the rice monocupins, subfamilies have members both with and without both UTRs, and thus it appears that there are members of each subfamily that may be expressed at low levels or not at all.
Only OsOXO4 was expressed in cvs. Vandana, Moroberekan or progenies between the two cultivars in the M. oryzae interaction. OsOXO4 was induced in both resistant and susceptible interactions, but the expression of OsOXO4 occurred 24 h earlier in the resistant interaction. In QTL-based defense response, it is common for resistance mechanisms to be expressed both in resistant and susceptible interactions with expression occurring sooner and more strongly with resistance (Bell et al. 1986; Chittoor et al. 1997; Steiner-Lange et al. 2003). Also, OsOXO4 expression was only detected after inoculation by the pathogen, suggesting that early induction of OsOXO4 expression, rather than basal expression, is important in defence responses to blast infection. This differs from the expression of the OsGLP on chromosome 8, which exhibited basal levels of expression prior to blast inoculation (Manosalva et al. 2008).
Expression of OsOXO4 can be correlated with EST data, where it is expressed in stressed leaves, as well as with MPSS data, where it is only expressed during the interactions with the rice blast and bacterial blight pathogens. Based on the MPSS data, expression of OsOXO4 was lower in the M. oryzae-Nipponbare (Pi-9) resistant interaction. However, our results show that it was induced at a higher level in the M. oryzae-Moroberekan resistant interaction. The lower level of expression in the MPSS data may be due to the resistance resulting from a major gene, Pi-9, which involves a different R gene-mediated resistance pathway (Qu et al. 2006).
A comparison of the 1,000-bp upstream of the transcriptional start site of the OsOXO4 in the cultivars Moroberekan and Vandana showed 100% identity, except for a 26-bp region that was not present in Vandana. In this region, four copies of the cis-elements related to bacterial nodulation responsive genes are found. This cis-element is similar to the VfLb29 promoter that was found to be specifically active not only in the infected cells of the nitrogen-fixing zone of root nodules but also in arbuscule-containing cells of transgenic Vicia faba and M. truncatula roots colonized by the endomycorrhizal fungus, Glomus intraradices (Vieweg et al. 2004). In Rhizobium–legume interactions, Nod-factor-induced changes in ion fluxes across membranes, followed by establishment of an oscillation of intracellular Ca2+ concentration, point to the involvement of a receptor-mediated signal transduction pathway (Downie and Walker 1999). This is similar to the oxidative burst involving OXO, where there is also a flux in intracellular Ca2+ concentration in the degradation of Ca-oxalate by OXO (Lamb and Dixon 1997). While both the susceptible and resistant rice varieties have the bacterial nodulation responsive cis-element in their promoters, the additional copies in Moroberekan and the resistant progenies may be responsible for the earlier expression of OsOXO4 compared to Vandana and the susceptible progeny. Similarly, in X. oryzae pv. oryzae–rice interaction, a slight difference in the promoter sequence of the resistance gene Xa27 also affected gene expression and disease resistance. For Xa27, the absence of a 10- and 25-bp region in the promoter of the allele from the resistant line was associated with resistance expression following inoculation with incompatible X. oryzae pv. oryzae strain (Gu et al. 2005).
The ratio of synonymous to nonsynonymous substitution was also examined among the monocupin subclasses. It is a measure of selection by comparing the rate of synonymous amino acid replacement with the rate of nonsynonymous amino acid replacements (Yang and Nielsen 2000). Among monocupins, this analysis suggests that the OXO and the OXLP subclasses are under purifying selection. Given the role of OXOs and OXLPs in disease resistance (Zhang et al. 1995; Hurkman and Tanaka 1996; Faris et al. 1999; Schweizer et al. 1999; Liu et al. 2004; Wu et al. 2004; Federico et al. 2006), one might have expected that those genes would all be under diversifying selection consistent with that of the NBS-LRR and other resistance (R) gene families (Richly et al. 2002; Meyers et al. 2003; Monosi et al. 2004; Fritz-Laylin et al. 2005; Mondragon-Palomino and Gaut 2005). However, R genes function in pathogen recognition and signal transduction to induce the HR, whereas OXOs and OXLPs are downstream of the signal transduction pathway for defense response (Tenhaken et al. 1995; Zhou et al. 1998) and may have different selection pressure than R genes. The barley GLPs, which have a role in defense response as well as plant development, are also under purifying selection (Federico et al. 2006). Thus, the purifying selection for the OXO and the OXLP subclasses could be consistent with the need for a particular structure/function in the defense response that favors preservation of protein function over coevolution with the pathogen.
Considerable evidence for the association of OXOs to resistance to fungal pathogens is available (Thompson et al. 1955; Zhou et al. 1998; Berna and Bernier 1999; Hu et al. 2003; Ramalingam et al. 2003; Livingstone et al. 2005). The barley HvOXO gene is expressed in the mesophyll tissue and the activity of the OXO enzyme increases in leaf extracts in response to Bgh, which may be a source of H2O2 (Zhou et al. 1998). Transgenic sunflower and peanuts transformed with wheat OXO and HvOXO, respectively, exhibited enhanced resistance to Sclerotinia species (Hu et al. 2003; Livingstone et al. 2005). OXOs were also mapped to QTLs involved in fungal resistance in wheat and rice (Faris et al. 1999; Ramalingam et al. 2003; Friesen and Faris 2004; Wu et al. 2004). The results of this study further support the role of specific OXO genes in resistance. The presence of OXOs only in cereals provides an interesting speculation into the evolutionary process in which members of the cupin superfamily diversified after the divergence of monocots and dicots 200 millions years ago (Wolfe et al. 1989). It is probably likely that specialization into OXOs occurred after the divergence.
Our study presents the rice OXOs as members of the cupin superfamily of proteins and highlights how analysis of gene structure can help identify candidates for contribution to disease resistance. OXOs are a unique group of proteins in cereals with converging evidences for their roles in defense response. We show that the OXOs form a cluster of four tandemly duplicated copies co-localizing with chromosome 3 QTL associated with resistance to rice blast. Specifically, we have shown that among the four tandemly duplicated OsOXOs, only OsOXO4 is expressed during M. oryzae–rice interaction, and it is expressed earlier during resistance than susceptibility. A unique feature of the resistance-associated OsOXO4 is that the resistance-associated sequence difference is not in the coding region but in a 26-bp sequence in the promoter region containing additional cis-elements associated with bacterial responsive nodulation genes. These cis-elements may also play a role in pathogen response. It would be interesting to know if transferring these cis-elements to the promoter of OsOXO4 in a susceptible cultivar, like Vandana, would also induce expression of OsOXO4 at an earlier time point and provide increased resistance.
Methods
Retrieval of cupin sequences and characterization of monocupin genes in the rice genome
To identify the complete set of proteins in the rice genome similar to OXO, we designed a PERL script to retrieve proteins with the cupin motif [G(X)5HXH-(X)3,4E(X)6G] and G(X)5PXG(X)2H(X)3N] from build 4 of the O. sativa ‘Nipponbare’ genome sequence obtained from the TIGR Rice Genome Annotation database (http://tigr.org/tdb/e2k1/osa1) (Yuan et al. 2005). Protein prediction was done using FGENESH (http://linux1.softberry.com/berry.phtml). ClustalX (Thompson et al. 1997) was used to align the protein sequences and generate phylogenetic trees. A total of 1,000 bootstrap replicates were performed using neighbor-joining, and clades having >60% bootstrap support were identified (Fritz-Laylin et al. 2005).
From the collection of cupin sequences, each sequence was classified into monocupin or bicupin using the Conserved Domain-Search option of GenBank’s Conserved Domain Database (Marchler-Bauer et al. 2005). The sequences were annotated by conducting BLAST search against the Swiss-Prot database for curated sequences (Schneider et al. 2004). The physical location of each sequence as well as the gene structure prediction were obtained from the TIGR Rice Annotation database and visualized by GenomePixelizer (Kozik et al. 2002).
ProtParam in Expasy (http://www.expassy.ch) was used to analyze the monocupin protein characteristics by determining amino acid length, isoelectric point stability index as well as hydrophobicity using Kyte-Doolitle method. To determine cellular localization, pSORT was used (http://wolfpsort.seq.cbrc.jp/). SignalP (http://www.cbs.dtu.dk/services/SignalP/) was employed to predict N-terminal signal peptides and MEME (Bailey and Gribskov 1998) was used to identify conserved protein motifs in the sequences. Selection pressure among monocupins was determined using codeml from the PAML package (Yang 1997; Yang and Nielsen 2000). A ω > 1 indicates diversifying selection while a ω < 1 indicates purifying selection. Of the six subclasses, only rhicadhesin-like receptor proteins, OXO and OXLP meet the criteria for calculating ω which are based on the following: (a) >50% similarity at the amino acid level, (b) absence of long insertions and deletions, and (c) at least three members. Evidence for monocupin gene expression was derived from the Rice Gene Expression Evidence page from TIGR (http://www.tigr.org/tdb/e2k1/osa1/locus_expression_evidence.shtml) and KOME (http://cdna01.dna.affrc.go.jp/cDNA/). The gene structure and orientation of monocupins were derived from the TIGR Rice Genome database build 5.0.
Expression of chromosome 3 OXOs
Plant materials used were O. sativa cvs. Moroberekan and Vandana and four advanced backcross progenies from a Vandana × Moroberekan cross (Wu et al. 2004). These materials were screened for resistance to rice blast under field conditions in India and the Philippines (Carrillo, Variar, Bhatt and Vera Cruz, unpublished). Molecular analysis of the progenies, IR78221-19-6-7-B, IR78221-19-6-56-B, and IR78221-19-6-99-B, show that they contain the QTL allele for OXO in chromosome 3 from resistant donor parent, Moroberekan, while the progeny, IR78221-19-3-196-B, contains the OXO allele from the susceptible parent, Vandana.
RT-PCR was used to detect the expression patterns of the OXO genes present on chromosome 3. Fifteen 14-day-old seedlings of each plant were inoculated with M. oryzae strain PO6-6 (100,000 spores/ml). Total RNA was extracted from leaves sampled at 0-, 6-, 12-, 24-, and 48-h post-inoculation using Trizol (Invitrogen, Carlsbad, CA, USA) and treated with DNAse (Promega, Madison, WI, USA). Two micrograms of RNA was used for first strand cDNA synthesis in a 20 µL reaction volume with Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s recommendations with oligodT(20) primer. Each cDNA was amplified using gene-specific OXO primer pair (10 μM) using 30 cycles of PCR amplification. Amplification of the actin gene, LOC_Os08g17820, was used as positive control for each gene. We designed gene-specific primers with the following sequences (5’–3’): (a) LOC_Os03g48750 (OsOXO1) F: ACCCTGACCCTCTCCAAGAC, R: CGGACCACCCTGGAGTAGTA; (b) LOC_Os03g48760 (OsOXO2) F: GGACCCTCTCCAGGACTTCT, R: CACGGACCACCTTGGAATAG; (c) LOC_Os03g48770 (OsOXO3) F: GCCTCTACGGGGTCTTGACT, R: CACGGACCACCTTGGAGTAG; (d) LOC_Os03g48780 (OsOXO4) F: AGCTTGTCACTGCGCTTCTT, R: GTGGCAATCTTGGAGGAGAA; and (e) Actin F: GTGGAGAGGCTGTGGAAGG, R: ATGTTCAGGGCTTTGCC.
In addition, the rice gene expression data from different expression libraries were also collected from the DCFI Rice Gene Index (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=Rice). The Rice Gene Index uses a highly refined protocol to analyze EST sequences in an attempt to identify the genes represented by that data and to provide additional information regarding those genes (Quackenbush et al. 2001). MPSS signatures for each OsOXO were derived from the Rice MPSS database (http://mpss.udel.edu/rice/). The Rice MPSS database is a repository of rice gene expression data derived from a sequencing-based technology that uses a unique method to quantify gene expression level, generating millions of short sequence tags per library (Meyers et al. 2004b; Nakano et al. 2006).
DNA isolation and sequencing
Leaves from 2-week-old greenhouse-grown plants were harvested for DNA extraction using the CTAB method. Gene-specific primers to amplify the four OXO genes in chromosome 3 were designed using the PRIMER3 program (Rozen and Skaletsky 2000). Three pairs of overlapping primers for sequencing the genes were designed that would encompass the 1-kb upstream region, the gene coding region and the 3’UTR region of each gene (as shown in Supplementary Table 5). The expected product sizes were 800–1,000 bp. DNA amplifications were performed in a 50-μL volume. The reactions contained 100 ng of genomic DNA, 0.25 mM of each primer, 2 mM of each dNTP, 1.50 U Taq DNA polymerase (New England Biolabs, Cambridge, MA, USA), and 1× PCR buffer (New England Biolabs, Cambridge, MA, USA). The amplification profile is as follows: 94°C for 5 min, 35 cycles of 1 min at 94°C, 1 min at 55°C, 2 min at 72°C, and followed by a final extension of 7 min at 72°C. PCR products were analyzed on agarose gel, purified using a Qiaquick PCR purification kit (Qiagen, Valencia, CA, USA), and quantitated prior to DNA sequencing. PCR products were sent to Macrogen (Seoul, Korea) for sequencing.
References
Bailey TL, Gribskov M. Methods and statistics for combining motif match scores. J Comput Biol 1998;5:211–21.
Bateman DF, Beer SV. Simultaneous production and synergitic action of oxalic acid and polygalacturonase during pathogenesis by Sclerotium rolfsii. Phytopathology 1965;55:204–11.
Bell JN, Ryder TB, Wingate VP, Bailey JA, Lamb CJ. Differential accumulation of plant defense gene transcripts in a compatible and an incompatible plant–pathogen interaction. Mol Cell Biol 1986;6:1615–23.
Berna A, Bernier F. Regulation by biotic and abiotic stress of a wheat germin gene encoding oxalate oxidase, a H2O2-producing enzyme. Plant Mol Biol 1999;39:539–49.
Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 2004;4:10.
Carter C, Thornburg RW. Tobacco nectarin I. Purification and characterization as a germin-like, manganese superoxide dismutase implicated in the defensE of floral reproductive tissues. J Biol Chem 2000;275:36726–33.
Carter C, Thornburg RW. The nectary-specific pattern of expression of the tobacco Nectarin I promoter is regulated by multiple promoter elements. Plant Mol Biol 2003;51:451–7.
Carter C, Graham RA, Thornburg RW. Arabidopsis thaliana contains a large family of germin-like proteins: characterization of cDNA and genomic sequences encoding 12 unique family members. Plant Mol Biol 1998;38:929.
Cessna SG, Sears VE, Dickman MB, Low PS. Oxalic acid, a pathogenicity factor for sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. The Plant Cell 2000;12:2191.
Chittoor JM, Leach JE, White FF. Differential induction of a peroxidase gene family during infection of rice by Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact 1997;10:861–71.
Doll J, Hause B, Demchenko K, Pawlowski K, Krajinski F. A member of the germin-like protein family is a highly conserved mycorrhiza-specific induced gene. Plant Cell Physiol 2003;44:1208–14.
Downie JA, Walker SA. Plant responses to nodulation factors. Current Op Plant Biol 1999;2:483–9.
Druka A, Kudrna D, Kannangara CG, von Wettstein D, Kleinhofs A. Physical and genetic mapping of barley (Hordeum vulgare) germin-like cDNAs. Proc Natl Acad Sci U S A 2002;99:850–5.
Dunwell J, Gibbings JG, Mahmood T, Naqvi S. Germin and germin-like proteins: evolution, structure, and function. Crit Rev Plant Sci 2008;27:342–75.
Dunwell JM, Khuri S, Gane PJ. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol Mol Biol Rev 2000;64:153–79.
Dunwell JM, Purvis A, Khuri S. Cupins: the most functionally diverse protein superfamily? Phytochemistry 2004;65:7–17.
Dunwell JM, Culham A, Carter CE, Sosa-Aguirre CR, Goodenough PW. Evolution of functional diversity in the cupin superfamily. Trends Biochem Sci 2001;26:740–6.
Elemento O, Gascuel O, Lefranc M-P. Reconstructing the duplication history of tandemly repeated genes. Mol Biol Evol 2002;19:278.
Faris JD, Li WL, Liu DJ, Chen PD, Gill BS. Candidate gene analysis of quantitative disease resistance in wheat. TAG Theor Appl Gen 1999;V98:219.
Federico ML, Iniguez-Luy FL, Skadsen RW, Kaeppler HF. Spatial and temporal divergence of expression in duplicated barley germin-like protein-encoding genes. Genetics 2006;174:179–90.
Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999;151:1531–45.
Friesen TL, Faris JD. Molecular mapping of resistance to Pyrenophora tritici-repentis race 5 and sensitivity to Ptr ToxB in wheat. TAG Theor Appl Gen 2004;109:464.
Fritz-Laylin LK, Krishnamurthy N, Tor M, Sjolander KV, Jones JD. Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol 2005;138:611–23.
Gane PJ, Dunwell JM, Warwicker J. Modeling based on the structure of vicilins predicts a histidine cluster in the active site of oxalate oxidase. J Mol Evol 1998;46:488–93.
Godoy G, Steadman JR, Dickman MB, Dam R. Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol Mol Plant Pathol 1990;37:179–91.
Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, et al. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 2005;435:1122–5.
Gucciardo S, Wisniewski JP, Brewin NJ, Bornemann S. A germin-like protein with superoxide dismutase activity in pea nodules with high protein sequence identity to a putative rhicadhesin receptor. J Exp Bot 2007;58:1161–71.
Guimaraes RL, Stotz HU. Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiol 2004;136:3703–11.
Hu X, Bidney DL, Yalpani N, Duvick JP, Crasta O, Folkerts O, et al. Overexpression of a gene encoding hydrogen peroxide-generating oxalate oxidase evokes defense responses in sunflower. Plant Physiol 2003;133:170.
Hurkman WJ, Tanaka CK. Germin gene expression is induced in wheat leaves by powdery mildew infection. Plant Physiol 1996;111:735–9.
IRGSP. The map-based sequence of the rice genome. Nature 2005;436:793–800.
Khuri S, Bakker FT, Dunwell JM. Phylogeny, function, and evolution of the cupins, a structurally conserved, functionally diverse superfamily of proteins. Mol Biol Evol 2001;18:593–605.
Kozik A, Kochetkova E, Michelmore R. GenomePixelizer—a visualization program for comparative genomics within and between species. Bioinformatics 2002;18:335.
Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 1997;48:251–75.
Lane BG. Oxalate, germin, and the extracellular matrix of higher plants. Faseb J 1994;8:294–301.
Lane BG, Dunwell JM, Ray JA, Schmitt MR, Cuming AC. Germin, a protein marker of early plant development, is an oxalate oxidase. J Biol Chem 1993;268:12239–42.
Liu B, Zhang S, Zhu X, Yang Q, Wu S, Mei M, et al. Candidate defense genes as predictors of quantitative blast resistance in rice. Mol Plant Microbe Interact 2004;17:1146–52.
Livingstone DM, Hampton JL, Phipps PM, Grabau EA. Enhancing resistance to Sclerotinia minor in peanut by expressing a barley oxalate oxidase gene. Plant Physiol 2005;137:1354–62.
Lynch M, O’Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics 2001;159:1789–804.
Manosalva PM, Davidson RM, Liu B, Zhu X, Hulbert SH, Leung H, Leach JE. A germin-like protein gene family functions as a complex QTL conferring broad-spectrum disease resistance in rice. Plant Physiol 2008;149:286–96.
Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, et al. CDD: a conserved domain database for protein classification. Nucleic Acids Res 2005;33:D192–196.
Membre N, Berna A, Neutelings G, David A, David H, Staiger D, et al. cDNA sequence, genomic organization and differential expression of three Arabidopsis genes for germin/oxalate oxidase-like proteins. Plant Mol Biol 1997;35:459–69.
Membre N, Bernier F, Staiger D, Berna A. Arabidopsis thaliana germin-like proteins: common and specific features point to a variety of functions. Planta 2000;211:345–54.
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 2003;15:809–34.
Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, et al. The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res 2004a;14:1641–53.
Meyers BC, Vu TH, Tej SS, Ghazal H, Matvienko M, Agrawal V, et al. Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat Biotechnol 2004b;22:1006–11.
Mondragon-Palomino M, Gaut BS. Gene conversion and the evolution of three leucine-rich repeat gene families in Arabidopsis thaliana. Mol Biol Evol 2005;22:2444.
Monosi B, Wisser RJ, Pennill L, Hulbert SH. Full-genome analysis of resistance gene homologues in rice. Theor Appl Genet 2004;109:1434–47.
Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucl Acids Res 2006;34:D731–735.
Ohno S. Evolution by gene duplication. New York: Springer; 1970.
Qu S, Liu G, Zhou B, Bellizzi M, Zeng L, Dai L, et al. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 2006;172:1901–14.
Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, et al. The TIGR gene indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 2001;29:159.
Ramalingam J, Vera Cruz CM, Kukreja K, Chittoor JM, Wu JL, Lee SW, et al. Candidate defense genes from rice, barley, and maize and their association with qualitative and quantitative resistance in rice. Mol Plant Microbe Interact 2003;16:14–24.
Richly E, Kurth J, Leister D. Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol 2002;19:76–84.
Rodriguez-Lopez M, Baroja-Fernandez E, Zandueta-Criado A, Moreno-Bruna B, Munoz FJ, Akazawa T, et al. Two isoforms of a nucleotide-sugar pyrophosphatase/phosphodiesterase from barley leaves (Hordeum vulgare L.) are distinct oligomers of HvGLP1, a germin-like protein. FEBS Lett 2001;490:44–8.
Roth C, Rastogi S, Arvestad L, Dittmar K, Light S, Ekman D, Liberles DA. Evolution after gene duplication: models, mechanisms, sequences, systems, and organisms. J Exp Zoolog B Mol Dev Evol. 2006.
Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 2000;132:365–86.
Schneider M, Tognolli M, Bairoch A. The Swiss-Prot protein knowledgebase and ExPASy: providing the plant community with high quality proteomic data and tools. Plant Physiol Biochem 2004;42:1013–21.
Schweizer P, Christoffel A, Dudler R. Transient expression of members of the germin-like gene family in epidermal cells of wheat confers disease resistance. Plant J 1999;20:541–52.
Steiner-Lange S, Fischer A, Boettcher A, Rouhara I, Liedgens H, Schmelzer E, et al. Differential defense reactions in leaf tissues of barley in response to infection by Rhynchosporium secalis and to treatment with a fungal avirulence gene product. Mol Plant Microbe Interact 2003;16:893–902.
Tenhaken R, Levine A, Brisson LF, Dixon RA, Lamb C. Function of the oxidative burst in hypersensitive disease resistance. Proc Natl Acad Sci U S A 1995;92:4158–63.
Thompson C, Dunwell JM, Johnstone CE, Lay V, Ray J, Schmitt M, et al. Degradation of oxalic acid by transgenic oilseed rape plants expressing oxalate oxidase. Euphytica 1955;85:169.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997;25:4876–82.
van Kan JAL. Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends in Plant Science 2006;11:247–53.
Vieweg MF, Fruhling M, Quandt HJ, Heim U, Baumlein H, Puhler A, et al. The promoter of the Vicia faba L. leghemoglobin gene VfLb29 is specifically activated in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots from different legume and nonlegume plants. Mol Plant-Microbe Interact 2004;17:62–9.
Wang GL, Mackill DJ, Bonman JM, McCouch SR, Champoux MC, Nelson RJ. RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar. Genetics 1994;136:1421–34.
Wei Y, Zhang Z, Andersen CH, Schmelzer E, Gregersen PL, Collinge DB, et al. An epidermis/papilla-specific oxalate oxidase-like protein in the defence response of barley attacked by the powdery mildew fungus. Plant Mol Biol 1998;36:101–12.
Wojtaszek P. Oxidative burst: an early plant response to pathogen infection. Biochem J 1997;322(Pt 3):681–92.
Wolfe KH, Gouy M, Yang YW, Sharp PM, Li WH. Date of the monocot–dicot divergence estimated from chloroplast DNA sequence data. Proc Nat Acad Sci U S A 1989;86:6201–5.
Wu JL, Sinha PK, Variar M, Zheng KL, Leach JE, Courtois B, et al. Association between molecular markers and blast resistance in an advanced backcross population of rice. Theor Appl Genet 2004;108:1024–32.
Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 1997;13:555–6.
Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol 2000;17:32.
Yang Z, Nielsen R, Hasegawa M. Models of amino acid substitution and applications to mitochondrial protein evolution. Mol Biol Evol 1998;15:1600.
Yuan Q, Ouyang S, Wang A, Zhu W, Maiti R, Lin H, et al. The institute for genomic research Osa1 rice genome annotation database. Plant Physiol 2005;138:18–26.
Zhang Z, Collinge DB, Thordal-Christensen H. Germin-like oxalate oxidase, a H2O2-producing enzyme, accumulates in barley attacked by the powdery mildew fungus. Plant J 1995;8:139.
Zhou F, Zhang Z, Gregersen PL, Mikkelsen JD, de Neergaard E, Collinge DB, Thordal-Christensen H. Molecular characterization of the oxalate oxidase involved in the response of barley to the powdery mildew fungus. Plant Physiol 1998;117:33–41.
Acknowledgment
We gratefully acknowledge the financial support from the Swiss Agency for Development and Cooperation and the Generation Challenge Program. We thank Kenneth McNally, Richard Bruskiewich, and Jun Ulat for advice on bioinformatics and Rebecca Davidson and Teresa de Leon for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
12284_2009_9024_MOESM1_ESM.ppt
Supplementary Fig. 1 Multiple sequence alignment of oxalate oxidases from the genome of the Nipponbare rice cultivar. a and b corresponds to the amino acid motif A and motif B of cupins, respectively. a Multiple sequence alignment of the nucleotide sequence of rice OXOs showing the 5’ UTR region. b Multiple sequence alignment of the amino acid sequences of the rice oxalate oxidases. A 23-amino acid signal peptide is found at the N terminus region of each gene as identified by pSORT (http://www.psort.org). (PPT 410 KB).
12284_2009_9024_MOESM2_ESM.ppt
Supplementary Fig. 2 Multiple sequence alignment of the predicted cupin domain of rice monocupin proteins. Motif A is [G(X)5HXH-(X)3,4E(X)6G] and Motif B is [G(X)5PXG(X)2H(X)3N]. The length of the intermotif region, located between the two motifs, is characteristic of particular genes. LOC_Os03g58990, which has a truncated protein sequence, and LOC_Os04g22080, which includes a retrotransposon-gag sequence in place of Domain B, are excluded from the alignment. (PPT 200 KB).
12284_2009_9024_MOESM3_ESM.ppt
Supplementary Fig. 3 A comparison of the gene structure of each monocupin gene. Arrangement is based on the phylogenetic tree in Figure 1. The orientation of the gene indicates its position with respect to the pseudomolecule (5’–3’). The exons are colored blue while the 5’ and 3’ UTR regions are white. Lines connecting two exons are the intron region. Genes with EST data available are boldfaced. Gene structures were derived from the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) (PPT 250 KB).
12284_2009_9024_MOESM4_ESM.ppt
Supplementary Fig. 4 Predicted protein sequence alight of OsOXO1 and OsOXO4. a Multiple alignment of OsOXO1 sequences from Moroberekan, Vandana, IR78221-19-6-7-B, IR78221-19-6-56-B, IR78221-19-6-99-B, and IR78221-19-3-196-B. A G-T replacement at position 136 changes the predicted amino acid from leucine to valine in Moroberekan and Vandana, respectively. b Pairwise alignment of OsOXO4 sequences from Moroberekan and Vandana. Arrows indicate amino acid substitutions in the predicted OsOXO4 protein. The resistant progenies IR78221-19-6-7-B, IR78221-19-6-56-B, and IR78221-19-6-99-B have the same sequence as Moroberekan while the susceptible progeny IR78221-19-3-196-B has the same sequence as Vandana. (PPT 1.70 MB).
12284_2009_9024_MOESM5_ESM.ppt
Supplementary Fig. 5 Multiple alignment of OsOXO2 sequences from Moroberekan, Vandana, IR78221-19-6-7-B, IR78221-19-6-56-B, IR78221-19-6-99-B, and IR78221-19-3-196-B. a A CG (red arrow) insertion in Vandana and the susceptible progeny IR78221-19-3-196-B in position 122 results to a stop codon in position 145. b Predicted amino acid sequence of OsOXO2. Blue arrow indicates a stop codon position resulting to a truncated protein in Vandana and IR78221-19-3-196-B. (PPT 1.58 MB).
12284_2009_9024_MOESM6_ESM.ppt
Supplementary Fig. 6 Abundance of OsOXO expressed sequence tags (ESTs) from The DFCI Oryza sativa Gene Index (OsGI). No ESTs were detected for OsOXO2. EST abundance of rice OxOs were retrieved from the Rice Gene Index (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb = Rice). OsOXO1 was expressed in the panicles and at flowering and pollination stages. OsOXO3 ESTs were found in roots and seed libraries. OsOXO4 ESTs were detected in drought, cold stress, and metal (CuSO4) stress libraries as well as in root, shoot, and leaf libraries. It was also expressed at tillering and booting stages. (PPT 84.5 KB).
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Carrillo, M.G.C., Goodwin, P.H., Leach, J.E. et al. Phylogenomic Relationships of Rice Oxalate Oxidases to the Cupin Superfamily and Their Association with Disease Resistance QTL. Rice 2, 67–79 (2009). https://doi.org/10.1007/s12284-009-9024-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12284-009-9024-0