Abstract
Mesencephalic astrocyte-derived neurotrophic factor (MANF) is an endoplasmic reticulum (ER) resident protein that can be secreted due to an imperfect KDEL motif. MANF plays a cytoprotective role in several soft tissues and is upregulated in conditions resulting from intracellular retention of mutant protein, including two skeletal diseases, metaphyseal chondrodysplasia, Schmid type (MCDS) and multiple epiphyseal dysplasia (MED). The role of MANF in skeletal tissue homeostasis is currently unknown. Interestingly, cartilage-specific deletion of Manf in a mouse model of MED resulted in increased disease severity, suggesting its upregulation may be chondroprotective. Treatment of MED chondrocytes with exogenous MANF led to a decrease in the cellular levels of BiP (GRP78), confirming MANF’s potential to modulate ER stress responses. However, it did not alleviate the intracellular retention of mutant matrilin-3, suggesting that it is the intracellular MANF that is of importance in the pathobiology of skeletal dysplasias. The Col2Cre-driven deletion of Manf from mouse cartilage resulted in a chondrodysplasia-like phenotype. Interestingly, ablation of MANF in cartilage did not have extracellular consequences but led to an upregulation of several ER-resident chaperones including BiP. This apparent induction of ER stress in turn led to dysregulated chondrocyte apoptosis and decreased proliferation, resulting in reduced long bone growth. We have previously shown that ER stress is an underlying disease mechanism for several skeletal dysplasias. The cartilage-specific deletion of Manf described in this study phenocopies our previously published chondrodysplasia models, further confirming that ER stress itself is sufficient to disrupt skeletal growth and thus represents a potential therapeutic target.
Similar content being viewed by others
Introduction
Mesencephalic astrocyte-derived neurotrophic factor (MANF) is an endoplasmic reticulum (ER)-stress associated protein that was initially identified as a neuroprotective factor in dopaminergic neurons (Hellman et al. 2011; Petrova et al. 2003). MANF has neuroprotective and cardioprotective effects, especially against ischemia-induced ER stress, and can promote cell proliferation in several tissues (Airavaara et al. 2009; Apostolou et al. 2008; Tadimalla et al. 2008). Mutations in MANF have been found in several tumours (Tanaka et al. 2000; Shridhar et al. 1997; Shridhar et al. 1996a, b), whilst increased expression of MANF has also been demonstrated in cytokine-induced ER stress (Cunha et al. 2017) and in diseases where the unfolded protein response (UPR) is induced by a misfolded mutant protein, such as in metaphyseal chondrodysplasia, Schmid type (MCDS; OMIM #156500 and resulting from mutations in COL10A1) or multiple epiphyseal dysplasia (MED; #607078 and resulting from mutations in MATN3) (Leighton et al. 2007; Nundlall et al. 2010; Cameron et al. 2011).
MANF is an ER-resident protein; however, it contains an imperfect KDEL sequence that allows it to be secreted under ER stress, suggesting it could be used as a biomarker for a subset of ER stress-related conditions (Henderson et al. 2013; Oh-Hashi et al. 2012). Moreover, exogenous MANF has been shown to have cytoprotective effects, potentially exerted via cell surface KDEL receptors (Henderson et al. 2013), and works in conjunction with the canonical ER chaperone BiP (GRP78). However, data on the effect of exogenous MANF on BiP expression suggests that this relationship might be cell or stress-type specific (Apostolou et al. 2008; Huang et al. 2016; Zhao et al. 2013).
Manf was identified as one of the most highly upregulated genes in the transcriptomic analysis of a mouse model of MED resulting from a mutation in the gene encoding matrilin-3, a structural molecule of the cartilage extracellular matrix (ECM) (Nundlall et al. 2010). MATN3-related MED results from canonical ER stress induced by the expression and misfolding of mutant matrilin-3 that aggregates over time in the ER lumen (Leighton et al. 2007; Nundlall et al. 2010). Manf was also upregulated in a related skeletal dysplasia, MCDS, which results from misfolding of type X collagen, a cartilage ECM protein exclusively expressed by hypertrophic chondrocytes of the growth plate (Hartley et al. 2013; Cameron et al. 2011).
MANF protein possesses several CXXC motifs that are shared amongst protein disulphide isomerases, suggesting a potential role in protein folding; however, we have previously shown that although MANF is present in the ER, it does not function as a protein disulphide isomerase (PDI) (Hartley et al. 2013).
The role of MANF in regulating the UPR, specifically during development of unchallenged healthy tissues, remains largely unknown. In this paper, we present data showing the importance of MANF in cartilage development and show that MANF exists in the chondrocyte ER in an interaction complex with several other ER resident chaperone proteins. Ablation of MANF from cartilage led to an imbalance of the UPR machinery and induced a protein kinase R (PKR)-like ER kinase (PERK)-mediated ER stress response that in turn resulted in decreased chondrocyte proliferation and reduced long bone growth. The results of this study further confirm that the induction of ER stress, whether by the expression of a mutant protein, or a genetically engineered deregulation of the UPR, results in a chondrodysplasia-like phenotype.
Methods
Generation of transgenic animals
Manf null mice were generated using targeted C57BL6 mouse embryonic stem cell clone Manftm1a(KOMP)Wtsi obtained from the KOMP Repository (http://www.komp.org). All the mice were generated on the C57BL6/J background to control for the genetic background effects. C57BL6 blastocysts were provided by the Animal Unit at University of Manchester, resulting in C57BL6 pure Manf null line (Manf−/−). Southern blotting was performed to confirm correct targeting (not shown), and Western blotting was performed on cartilage to confirm deletion of MANF in mutant chondrocytes. Manf conditional line (Manffl/fl Col2Cre+) was generated by restoring the wild-type allele through crossing the Manf−/− mice with an Actin-Flp (C57BL6) mouse and then by crossing the mice with a C57BL6 Col2Cre expressing line (Sakai et al. 2001). To analyse the expression pattern of Col2cre, mice expressing the Col2Cre transgene were crossed with mice homozygous for the Gtrosa26tm1Sor-targeted mutation consisting of a bacterial β-galactosidase gene flanked by two loxP sites. All experiments were performed in compliance with the Scientific Procedures Act of 1986 and the relevant Home Office (under PPL 40/2884 and PPL60/04525) and Institutional regulations governing animal breeding and handling.
LacZ staining of embryos and tissues
Gtrosa26tm1Sor mice were bred with a Cre expressing strain, which resulted in removal of a DNA fragment that prevents transcription of the lacZ gene in the tissues expressing Cre recombinase. Tissue expression pattern of the Cre transgene was then assayed using X-gal staining. Gtrosa26tm1Sor;Col2Cre+ and Gtrosa26tm1Sor;Col2Cre− embryos at embryonic day 12.5 were fixed for 15 min in 2.0% formaldehyde and 0.2% glutaraldehyde. Postnatal 1-week old limbs from Gtrosa26tm1Sor;Col2Cre+ and Gtrosa26tm1Sor;Col2Cre− mice were harvested and fixed in 10% neutral buffered formalin for 4 h at room temperature. The samples were washed in 0.01% sodium deoxycholate and 0.02% Nonidet™ P-40 and placed in lacZ staining solution containing 1 mg/ml X-Gal overnight in a humidified darkened chamber at 37 °C. Following a wash in 1× PBS, the embryos were fixed in 10% neutral buffered formalin overnight and incubated in 1% formaldehyde solution for long-term storage. The limbs were decalcified by gentle agitation in 20% EDTA pH 7.4, embedded in OCT medium and sectioned at 10-μm sections using Leica CM1860 Cryostat. Sections were counterstained with eosin, dehydrated through a series of ethanol concentrations and mounted in HistoMount™.
Bone measurements
X-rays were acquired using the MX-20 Cabinet X-ray System (Faxitron® Bioptics) under transient anaesthesia, allowing for longitudinal bone growth measurements at 3 and 9 weeks of age. Bones were measured using ImageJ (National Institutes of Health, Bethesda, MD, USA; Rueden et al. 2017). One-way ANOVA was performed using GraphPad Prism version 7.0 (GraphPad Software, La Jolla, CA, USA, www.graphpad.com) for statistical analysis of the data.
Histology and immunohistochemistry
Embryos were retrieved by caesarean section at E18.5, and their lungs were collected. One-week-old mice and adult mice were culled by anaesthetic overdose, and hind limbs were harvested. Hind limbs and lungs were fixed in either 4% paraformaldehyde (PFA, for histology) or 95% ethanol 5% acetic acid (immunohistochemistry) for 48 h at 4 °C. The limbs were then decalcified in 20% EDTA pH 7.4 for 2 weeks, embedded in paraffin and cut into 6-μm sections. Haematoxylin/eosin (H&E) staining was performed to visualise the general morphology of the tissue, using the Thermo Scientific™ Linistat™ Slide Stainer. To analyse the expression pattern of the Col2cre transgene in the limbs, 1-week-old PFA-fixed sections were stained overnight in a LacZ staining solution containing 1 mg/mL X-Gal. Immunohistochemistry and bromodeoxyuridine (BrdU) labelling were performed as described previously (Pirog-Garcia et al. 2007) using the appropriate Alexa Fluor® secondary antibodies. For immunohistochemistry, slides were mounted in Fluoroshield™ Mounting Media with DAPI (Abcam®). Primary antibodies were used at a dilution of 1:500 (type II collagen (ab34712, Abcam®); Matrilin-3 (AF3357, R&D Systems®; type X collagen); Rajpar et al. 2009). Images were obtained using the Zeiss Axio Imager 2 microscope. BrdU-labelled cells were counted using the watershed algorithm on the Fiji ImageJ platform (National Institutes of Health, Bethesda, MD, USA; Schindelin et al. 2012) and presented as percentage of total cells in the proliferative zone.
TUNEL assay
TUNEL assay was used to analyse cell death on 4% PFA fixed sections of 3-week-old limbs using the Promega® DeadEnd™ Fluorometric TUNEL System according to the manufacturer’s protocol. Antigen unmasking was performed using a citric buffer boil pH 6 boil instead of proteinase K unmasking that can generate false positives (Gál et al. 2000; Pirog-Garcia et al. 2007). Sections were mounted in Fluoroshield™ mounting media with DAPI, and images were obtained using the Zeiss Axio Imager 2 microscope. Positive TUNEL cells were counted using the watershed algorithm on Fiji ImageJ platform (National Institutes of Health, Bethesda, MD, USA; Schindelin et al. 2012) and presented as percentage of all cells in selected zones of the growth plate. One-way ANOVA was performed using GraphPad Prism version 7.0 (GraphPad Software, La Jolla CA, USA, www.graphpad.com) for statistical analysis of the data.
RNA extraction and RNA-sequencing analysis of knee chondrocytes
Whole knee joints from 5-day-old mice were dissected and cleaned free of soft tissue following digestion for 1 h in 2.5 mg/mL collagenase IA. Joints were homogenised using a Satorius Mikro-Dismembranator S, and RNA was extracted using the Promega® ReliaPrep™ RNA Tissue Miniprep System according to manufacturer’s protocol.
The extracted RNA was sent to GATC Biotech for analysis. Each sample contained RNA extracted from three littermates, pooled into one sample at a concentration of 1 μg total RNA with a RNA integrity number (RIN) ≥ 8. The RNA was sequenced and analysed by GATC Biotech. Briefly, RNA sequencing reads were aligned to the mouse reference genome using Bowtie. Top Hat was used to identify exon–exon splice junctions of the initial alignment. Cufflinks and Cuffmerge were used to identify, quantify, merge and annotate the transcripts from the processed RNA-Seq alignment assembly. The merged transcripts from the wild type (Manffl/fl Col2Cre−) and Manffl/fl Col2Cre+ samples were compared using Cuffdiff to determine differential expression levels with a measure of significance between the samples.
Extraction and treatment of primary chondrocytes
Costal and tibial chondrocytes were isolated from pooled litters of 5-day-old mice (Nundlall et al. 2010) and cultured for up to a week in DMEM/F12 GlutaMAX™ supplement medium (Fisher Scientific™) containing 5% FBS, 5% non-essential amino acids, 1 U/mL penicillin, 1 μg/mL streptomycin and 50 g/mL l-ascorbate-2-phosphate.
Once confluent, the extracted cells were treated with 1 mM thapsigargin or 1 μg/mL tunicamycin in DMSO to induce ER stress. After 4 h, 100 ng/mL recombinant human MANF (3748-MN-050, R&D Systems®) was added to the media and cells were cultured for a further 24 h. To analyse the effect of MANF on primary V194D matrilin-3 chondrocytes, 100 ng/mL recombinant human MANF was added to media and cells were cultured for 24 h.
Protein extraction and immunoblotting
Protein lysates were prepared from cells in monolayer in 1× RIPA buffer (50 mM Tris–HCl, pH 7.4, 1% Triton X-100, 150 mM NaCl, 1 mM EDTA) and centrifugation at 13,600×g. Samples were denatured by boiling at 95 °C for 5 min in SDS-PAGE loading buffer containing 100 mM dithiothreitol (DTT) for reducing conditions. Proteins were separated according to size by SDS-PAGE using precast Novex™ NuPAGE® 4–12% Bis-Tris precast gels (Fisher Scientific™) in MES SDS-PAGE running buffer (Fisher Scientific™) at 200 V for 60 min. Proteins were then electroblotted onto a nitrocellulose membrane for 1 h at 30 V using the XCell II™ Blot Module (Fisher Scientific™). Gel loading was assessed using REVERT™ Total Protein Stain (LI-COR® Biosciences). The membrane was then destained using REVERT™ Reversal Solution according to manufacturer’s instructions. Membranes were washed and blocked in 3% milk in PBS-T then incubated with primary antibodies for 1 h at room temperature. Primary antibodies used were GRP78 (ab108615, Abcam®), GRP94 (sc-1794, Santa Cruz Biotechnology), PDIA6 (ab154820, Abcam®) and matrilin-3 (AF3357, R&D). Membranes were then probed with the appropriate LI-COR® IRDye® secondary antibody at a concentration of 1:10,000 for 1 h. Blots were imaged on the LI-COR® Odyssey CLx Imaging System, and band intensity was normalised to the total protein stain using the LI-COR® proprietary software. Loading control images (total protein) are included in Online Resource 1, Fig. 3. Student’s t test was performed using GraphPad Prism version 7.0 (GraphPad Software, La Jolla, CA, USA, www.graphpad.com) for statistical analysis of the data.
Co-immunoprecipitation
Co-immunoprecipitation (Co-IP) was carried out on cell lysates from HEK-293 cells transfected with FLAG-tagged and V5-tagged wild-type MANF constructs in pcDNA3.1+ using anti-FLAG and anti-V5 agarose beads (ab1299, Abcam®). Prior to immunoprecipitation, cells were treated with 1 mM dithiobis[succinimidyl propionate] (DSP) to stabilise protein complexes as previously described (Cotterill et al. 2005). Cell lysates were prepared as outlined above. Co-IP was performed using 500 μg of cell lysate, which was incubated with anti-V5 agarose beads overnight under gentle agitation as per manufacturer’s instructions. The resin was pelleted by centrifugation and washed three times in 1× RIPA buffer. Proteins were eluted in SDS buffer, and SDS-PAGE was performed as above.
Mass spectrometry and data analysis
Twenty-microliter aliquots of cross-linked Co-IP cell-lysates were run into 4–12% SDS-polyacrylamide gels for 4 min (at 200 V). Total protein pools were excised from the gel, dehydrated, reduced, alkylated and washed. Samples were then digested with trypsin overnight at 37 °C and analysed by LC-MS/MS. Peptides were concentrated on a precolumn (C18 PepMap, 20 mm × 180-μm i.d, Thermo) and separated using a gradient from 95% A (0.1% formic acid in 2% acetonitrile) and 5% B (0.1% formic acid in acetonitrile) to 35% B, in 132 min at 300 nL min−1, using a 75-μm × 250-mm i.d. 3-μm particle size PepMap C18, analytical column (Waters) on an Ultimate 3000 nano-HPLC system (Thermo) coupled to an LTQ Orbitrap XL mass spectrometer (Thermo). The capillary voltage was set to 200 °C and the spray voltage to 1.6 kV. Survey scans were acquired in the Orbitrap with a resolution of 30,000 at m/z 400 Da. Up to 15 data-dependent MSMS scans were acquired in the LTQ following CID. Peak lists were generated using msconvert, and the data was searched using X!Tandem and the GPM interface against a concatenation of the ENSEMBL Homo sapiens genome (v. 76) and cRAP (v.2012.01.01) (Kessner et al. 2008; Craig et al. 2004). The following search parameters were used: tryptic cleavage; precursor mass accuracy 100 ppm; product ion mass tolerance 0.6 Da; static modification- carbamidomethylation on C; variable modification- oxidation on Met; refinement- yes; refinement modifications- Deamidated@N or Q, Phospho@S or T or Y, Oxidation@M or W, Methyl@C or D or E or H or R or K or N or Q, Dioxidation@M or W, Dehydrated@S or T, -Carbamidomethyl@C, Carbamidomethyl@H or D or E or K. Proteins were considered to be identified when they had protein level log(e) values < (− 3). Proteins present in cRAP were removed from the result lists. Three biological replicates were used in all experiments. The number of spectra identified for each protein was compared between wild-type and mutant genotypes using the Student’s t test GraphPad Software, La Jolla, CA, USA, www.graphpad.com) with a P value < 0.05 was considered significant.
Results
Global deletion of Manf leads to perinatal lethality due to breathing difficulties induced by lung malformation
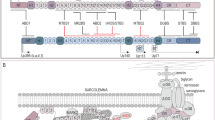
Manf null mice were generated using targeted C57BL/6 mouse embryonic stem cell clones obtained from the KOMP Repository (http://www.komp.org,). The clones were injected into C57BL/6 blastocysts resulting in a Manf null line (Manf−/−; Fig. 1a). The lack of MANF expression was confirmed by Western blotting in liver tissue samples (Fig. 1b). Interestingly, heterozygous breeding pairs (Manf+/− × Manf+/−) failed to produce Manf null offspring, with a genotype ratio of Manf+/−:Manf+/−:Manf−/− offspring of 33%:67%:0% over 14 litters from 3 different breeding pairs. Embryos were therefore recovered at E18.5 by caesarean section in an attempt to foster; however, they failed to breathe when induced.
a Targeting strategy used to generate the Manf−/− (‘knockout first’) and Manffl/fl Col2Cre+ (conditional knockout allele) lines. Black boxes represent Manf exons; LoxP sites are marked by arrowheads and FRT sites by ovals. b Western blotting showing the deletion of MANF in Manf−/− liver at E19.5, GAPDH as loading control. c LacZ staining of the whole mount ß-galactosidase (Rosa26) reporter crossed with Col2Cre line embryo at E15.5, showing cartilage-specific expression of the Col2Cre transgene. d Western blotting showing deletion of MANF in Manffl/fl Col2Cre+ cartilage at P21, GAPDH as loading control. Scale bars 1 mm and 100 μm
In order to understand the underlying pathology of these mice, the lungs were collected from embryos at E17.5, E18.5 and E19.5 days to analyse the respiratory capacity of the animals and to identify any potential respiratory impairment. Following haematoxylin and eosin of lung sections, the alveolar volume was measured using ImageJ. The alveolar volume was significantly decreased in the Manf null mice at all ages analysed, thus confirming a role for Manf in embryonic lung development (Online Resource 1, Fig. 1a, b).
Endochondral ossification is impaired in cartilage-specific Manf knockout mice
Based on our previous studies (Piróg et al. 2014), and other published evidence (Dixon and Dixon 2004; McLaughlin et al. 2007), we have hypothesised that a C57BL/6 genetic background can exaggerate the skeletal disease phenotype in ER stress-related mouse models. Indeed, crossing the global Manf null mice onto a 129Sv background (50%:50% C57BL/6:129Sv) resulted in viable offspring, which is consistent with a previously published study (Lindahl et al. 2014). However, global Manf null mice were much smaller than their littermates and later became diabetic (Online Resource 1, Fig. 2a). In order to eliminate systemic and metabolic effects on bone growth, and to study the specific role of Manf in cartilage development and homeostasis, we generated a conditional knockout mouse line. Manf null animals were crossed with a Flp recombinase strain to restore a wild-type allele and then with a Col2Cre expressing mouse (Gualeni et al. 2013) (Fig. 1c) in order to delete exon 3 of Manf in cartilage only (Fig. 1a). SDS-PAGE and Western blotting were performed on femoral head cartilage dissected from 5-day-old mice to confirm that MANF was absent in Manffl/fl Col2Cre+ mice (Fig. 1d).
Manffl/fl Col2Cre+ mice had shorter long bones at both 3 weeks (not shown) and 9 weeks of age (Fig. 2a, c; tibia ↓5.4% P < 0.001; femur ↓3.5% P < 0.001) and reduced skull lengths (measured from the tip of the nose to the base of the skull (Fig. 2a, c; skull ↓4.5% P < 0.05)), indicating a disruption to endochondral ossification. Interestingly, the Manf null cartilage growth plate appeared morphologically normal and showed the typical columnar organisation of chondrocytes that was comparable to wild-type mice (Fig. 2b; Online Resource 1, Fig. 2b). Moreover, the location of key cartilage ECM components such as types II and X collagen and matrilin-3 was unchanged in Manffl/fl Col2Cre+ growth plates, thus indicating that MANF is not essential for the processing and secretion of these key structural components of the cartilage growth plate (Fig. 3a).
X-ray radiographs of Manffl/fl Col2Cre− and Col2Cre+ mice at 9 weeks. b Haematoxylin and eosin staining of Manffl/fl Col2Cre− and Col2Cre+ cartilage growth plates at P21. c Bone measurements of Manffl/fl Col2Cre− and Col2Cre+ mice at P63 showing affected endochondral ossification elements (n > 5). RZ resting zone, PZ proliferative zone, HZ hypertrophic zone. *P < 0.05, ***P < 0.0001. Scale bars 5 mm (a) and 200 μm (b)
a Immunohistochemistry showing the localisation of cartilage extracellular matrix components, type II collagen, type X collagen and matrilin-3 in Manffl/fl Col2Cre− and Col2Cre+ growth plates at P21. b 2 h BrdU labelling of proliferation in Manffl/fl Col2Cre− and Col2Cre+ growth plates at P21 showing a 29% decrease in chondrocyte proliferation upon deletion of MANF (n = 5). c TUNEL assay per zone in Manffl/fl Col2Cre− and Col2Cre+ growth plates at P21 (n = 3). d Total apoptosis in Manffl/fl Col2Cre− and Col2Cre+ growth plates at P21 (n = 3). RZ resting zone, PZ proliferative zone, HZ hypertrophic zone, NC negative control. **P < 0.01. Scale bar 200 μm
Deletion of MANF from the cartilage growth plate leads to a decrease in chondrocyte proliferation and dysregulated apoptosis
A 2-h in vivo labelling by BrdU was used to measure chondrocyte proliferation in Manf wild type (Manffl/fl Col2Cre−) and Manffl/fl Col2Cre+ growth plate cartilage at 3 weeks of age. We measured a 29% decrease in chondrocyte proliferation following MANF deletion suggesting an underlying cellular stress (Fig. 3b; P < 0.01, n = 5,).
The TUNEL assay was used to identify apoptosis in wild type (Manffl/fl Col2Cre−) and Manffl/fl Col2Cre+ growth plate cartilage at 3 weeks, and TUNEL positive cells were individually quantified in the resting, proliferative and hypertrophic zones respectively. We detected no differences in chondrocyte apoptosis in the resting or hypertrophic zones of Manffl/fl Col2Cre+ growth plate cartilage when compared to the wild-type (Manffl/fl Col2Cre−) littermates, but a 2.5-fold increase in TUNEL positive cells in the proliferative zone (Fig. 3c; n = 3). However, the overall proportion of TUNEL positive cells in the total growth plate was similar between the Manffl/fl Col2Cre− and Manffl/fl Col2Cre+ mice, suggesting premature (stress-induced) apoptosis was occurring in chondrocytes of the proliferating zone (Fig. 3d).
The ablation of MANF in growth plate cartilage leads to an increased ER stress response
Transcriptomic analysis (RNAseq) was performed on RNA extracted from the chondrocytes of the tibial cartilage of 5-day-old mice. Surprisingly, the expression of only seven genes was differentially changed between wild type (Manffl/fl Col2Cre−) and Manffl/fl Col2Cre+ chondrocytes (Table 1; P < 0.05 and a log2 fold change > 0.6).
The only downregulated gene was Manf itself, thus confirming its cartilage-specific ablation. The six upregulated genes were Chac1 (1.9-fold P = 7E−16), Trib3 (1.6-fold, P = 6.6E−08), Hyou1 (1.6-fold, P = 2.0E−18), Asns (1.6-fold, P = 1.2E−12), Hspa5 (1.5-fold, P = 7.1E−16) and Sdf2l1 (1.5-fold, P = 1.6E−06). Interestingly, all these genes encode ER-resident proteins that exist as a complex inside the ER lumen, including the sentinel ER stress response marker GRP78 (Hspa5) (Fig. 4a). When the data were reanalysed at a less stringent 1.2-fold cutoff level, several other ER-related genes (such as Atf5, Canx, Creld2, Dnajb9, Dnajc3, Edem1 and Xbp1; Online Resource 2) were upregulated, whilst downregulated genes pertained mostly to cell adhesion and differentiation. Moreover, Ihh was upregulated 1.3-fold (P = 0.03), potentially reflecting the dysregulated chondrocyte proliferation and differentiation. To validate the RNAseq, we confirmed that GRP78 protein (but not the related GRP94) was increased in cartilage from 5-day-old mice cartilage (Fig. 4b; 33% P > 0.05, n = 3).
a STRING network showing the known interactions between the proteins encoded by the genes differentially expressed in the Manffl/fl Col2Cre− and Col2Cre+ cartilage at P5. b Deletion of MANF increases the levels of GRP78 but not GRP94 in cartilage at P5, as shown by Western blotting and densitometry measurement (n = 3). c Western blotting of proteins co-immunoprecipitated in the 293 cells expressing FLAG-tagged recombinant MANF
MANF directly interacts with proteins involved in the unfolded protein response
After demonstrating that a specific subset of ER stress response genes was upregulated in MANF-null chondrocytes, we next investigated if these proteins existed as a complex with MANF. Co-immunoprecipitation was performed by overexpressing FLAG-tagged MANF in 293 cells and identifying protein complexes using mass spectrometry.
MANF co-immunoprecipitated with several ER components including CCT5, GRP78, PDIA6, PRDX4 and TCP1 (Table 2; Fig. 4c; full MS analysis in Online Resource 3). MANF also bounds several ribosomal components further corroborating its role in protein synthesis and folding (Hartley et al. 2013). GO term analysis of the proteins associated with MANF clustered them into two categories: ‘protein processing in the ER’ and ‘ribosome’.
Exogenous MANF rescues primary chondrocytes from tunicamycin-induced ER stress
Exogenous MANF has been shown to alleviate ER stress in pancreas, brain and heart tissues (Apostolou et al. 2008; Huang et al. 2016; Zhao et al. 2013). To explore if there is a similar role for MANF in cartilage, primary chondrocytes were extracted from wild-type costochondral cartilage and treated with 1 μg/mL tunicamycin to induce ER stress by blocking N-linked glycosylation. The cells were labelled with BrdU for 2 h to determine the effect on cell proliferation, and not surprisingly, tunicamycin treatment significantly reduced chondrocyte proliferation (Fig. 5a: by 64%, P > 0.05). The addition of exogenous MANF had no significant effect on the proliferation of untreated chondrocytes; however, it led to a 19% increase in the proliferation of tunicamycin-treated chondrocytes, suggesting that it may have alleviated to some extent the tunicamycin-induced ER stress (Fig. 5a). The influence of MANF on tunicamycin-induced ER stress was further confirmed by immunoblotting for GRP78. Levels of GRP78 increased by 46% following tunicamycin treatment, indicating high levels of ER stress, but were reduced by 14% following treatment with exogenous MANF (Fig. 5b, P < 0.01 and P < 0.05, respectively). Overall, these data suggested the potential for exogenous MANF to reduce ER stress.
a BrdU labelling of control primary chondrocytes and chondrocytes treated with tunicamycin with and without exogenous MANF (n = 2). b Western blot and densitometry measurement of GRP78 levels in untreated and tunicamycin-treated primary chondrocytes with and without exogenous MANF (n = 2). Tun tunicamycin. ***P < 0.001, **P < 0.01, *P < 0.05
MANF plays an important role in modulating the pathobiology of MED
After demonstrating that the deletion of Manf in mouse chondrocytes produced a growth plate dysplasia and chondrodysplasia-like phenotype, we next investigated its putative protective role in Matn3 MED chondrocytes. Manffl/fl Col2Cre+ mice were crossed with the Matn3V194D mouse model of MED (Leighton et al. 2007) to determine whether the upregulation of Manf expression in Matn3V194D chondrocytes (Nundlall et al. 2010) was a detrimental consequence or in fact beneficial and represented an attempt by the cells to alleviate the ER stress.
Mice that were homozygous for both mutant alleles (Manffl/fl and Matn3V194D) and carrying a copy of Col2Cre were dramatically shorter than their wild-type littermates and had breathing difficulties due to restricted rib cages. This obvious increase in disease severity confirmed a chondroprotective role for MANF in the pathobiology of MED (Fig. 6a). However, in vitro treatment of Matn3V194D chondrocytes with exogenous MANF had no effect on the amount or localisation of retained mutant matrilin-3 despite lowering the levels of GRP78 by 25% (Fig. 6b–d). Overall, these data suggested that intracellular MANF is of more importance to pathobiology of MED.
a X-ray radiographs of Manffl/fl Col2Cre− Matn3+/+, Manffl/fl Col2Cre− Matn3V194D/V194D and Manffl/fl Col2Cre+ Matn3V194D/V194D mice at 6 weeks, showing the dramatic effect of removal of MANF from cartilage of the mouse model of MED. b Western blot and densitometry measurement of intracellular GRP78 levels in Matn3V194D/V194D primary chondrocytes with and without exogenous MANF (n = 4). c Western blot and densitometry measurement of intracellular matrilin-3 levels in Matn3V194D/V194D primary chondrocytes with and without exogenous MANF (n = 4). d Immunocytochemistry for matrilin-3 showing that exogenous MANF has no effect on the intracellular retention of mutant matrilin-3 (in green) in Matn3V194D/V194D primary chondrocytes. DAPI was used as a counterstain. Scale bars 5 mm (a) and 200 μm (d) (colour figure online)
Discussion
MANF, also known as ARMET (arginine-rich mutated in early stage tumours), is a small arginine-rich protein found in the ER (Hartley et al. 2013; Mizobuchi et al. 2007). MANF is strongly expressed in the lung epithelium, liver, pancreas and brain (Shridhar et al. 1996a, b) and is cytoprotective against ischemia-induced ER stress in several tissues (Airavaara et al. 2009; Apostolou et al. 2008; Tadimalla et al. 2008). Following ER stress, MANF can be secreted from cells due to an imperfect KDEL motif at the C-terminus (Henderson et al. 2013; Oh-Hashi et al. 2012). It has been suggested that it is through the action of cell surface KDEL receptors and a putative novel receptor that exogenous MANF exerts its cytoprotective effect (Henderson et al. 2013). MANF has a similar expression pattern to BiP (GRP78) in mouse tissues (Mizobuchi et al. 2007), and BiP inhibition blocks exogenous MANF-mediated cell survival (Huang et al. 2016). The cytoprotective effect of MANF has been shown in several tissues, but the exact mechanism remains to be elucidated with some studies showing an increase (Huang et al. 2016), whilst others report a decrease in BiP upon MANF treatment (Zhao et al. 2013).
The study of a Manf null mouse model (in C57BL/6 × ICR mice) has previously been published in which the mice were viable, but over time, they developed diabetes and showed signs of pancreatic ER stress (Lindahl et al. 2014). In order to study the role of MANF in cartilage health and disease, we initially tried to generate a global Manf knockout mouse line on a pure C57BL/6 background; however, somewhat surprisingly, hemizygous C57BL/6 Manf+/− mice failed to produce viable Manf null offspring. Indeed, Manf null pups died perinatally through respiratory arrest due to a lung malformation characterised by decreased alveolar space and volume of individual alveoli. Interestingly, BiP (GRP78) has been shown to regulate distal epithelial cell survival during lung development and lungs from GRP78 null mice exhibit dilated alveolar airspaces and alveolar hypoplasia leading to perinatal respiratory arrest (Flodby et al. 2016). Deletion of Manf induces Hspa5 expression (Lindahl et al. 2014 and in this study) and the phenotype of the Manf null lungs are directly opposite to the phenotype seen following deletion of Hspa5 (Flodby et al. 2016), indicating that the GRP78/MANF relationship is important for lung tissue development.
MANF was previously identified as the most highly upregulated gene in a mouse model of MED resulting from a mutation in matrilin-3 (Nundlall et al. 2010). However, the role of MANF in cartilage and in the pathobiology of chondrodysplasias remains largely unknown. In order to study the specific effects of Manf ablation in cartilage, we generated a cartilage-specific knockout strain using Cre recombinase expressed under the type II collagen promoter (Sakai et al. 2001). The resultant conditional knockout mice had shorter bones, but the cartilage growth plate appeared morphologically normal with no overt ECM disruption. Interestingly, chondrocyte proliferation was significantly decreased in the Manf null cartilage when compared to the wild-type controls and apoptosis, whilst not increased overall, was dysregulated. We have previously shown that a reduction in chondrocyte proliferation is a key mechanistic finding in mouse models of chondrodysplasia caused by mutations in Matn3 (V194D ↓16%; Leighton et al. 2007) and COMP (T585M ↓24% (Pirog-Garcia et al. 2007) and 469Del ↓17% (Suleman et al. 2011)) and confirmed that reduced proliferation is the major driver of reduced long bone growth in mouse ER stress phenocopies (ColIITgrdw ↓21% (Gualeni et al. 2013) and ColIITgcog ↓12% (Rajpar et al. 2009)).
Transcriptomic analysis of chondrocytes from postnatal day 5 Manffl/fl Col2Cre+ cartilage revealed increased expression of only six genes whose protein products are all localised to the ER. These were the following: Chac1 (ChaC glutathione-specific gamma-glutamylcyclotransferase 1), an ER stress-related apoptosis and oxidative stress modulator downstream of ATF4 (Mungrue et al. 2009; Crawford et al. 2015); Trib3 (Tribbles pseudokinase 3), involved in modulation of CHOP-dependent cell death during ER stress (Fang et al. 2014; Qian et al. 2008) and decrease of proliferation of bone marrow-derived mesenchymal stem cells (Zhang et al. 2017); Hyou1 (hypoxia upregulated 1), shown to suppress hypoxia and CHOP-induced apoptosis (Wu et al. 2013; Ozawa et al. 1999); Asns (asparagine synthetase), an antiapoptotic molecule localised downstream of ATF4 in cancer and ER stress (Gjymishka et al. 2009; Balasubramanian et al. 2013); Hspa5 (the gene for GRP78), an upregulation of which inhibits CHOP-mediated apoptosis (Xiong et al. 2015); and Sdf2l1 (stromal cell-derived factor 2 like 1), an ER stress inducible gene interacting with GRP78 and modulating the activity of the ERAD system (Tiwari et al. 2013; Fujimori et al. 2017; Fukuda et al. 2001). In order to verify the RNAseq results, and to look for MANF-associated protein complexes, we performed co-immunoprecipitation by overexpressing FLAG-tagged recombinant MANF in 293 cells. Interestingly, MANF co-immunoprecipitated with GRP78, PRDX4 and PDIA6 amongst others, further confirming MANF’s involvement in ER chaperone complexes. Interestingly, all of the protein products of the genes upregulated in the absence of Manf are linked to the PERK arm of the UPR and modulation of ER stress-related apoptosis and have been shown to interact with each other (Zhang et al. 2002; Fels and Koumenis 2006; Kang et al. 2017; Meunier et al. 2002). Moreover, this finding is in agreement with data published for the Manf null pancreas, which also showed a specific upregulation of the PERK arm of the UPR pathway including GRP78, ATF4, PERK and CHOP (Lindahl et al. 2014).
Overall, these data suggest that the deletion of Manf introduces an imbalance in the chondrocyte ER homeostasis that in itself is sufficient to result in a chondrodysplasia-like phenotype. Chondrocytes in developing cartilage are highly secretory cells, producing a proteoglycan and a collagen-rich ECM, and exist in a state of mild physiological ER stress (Hughes et al. 2017; Hino et al. 2014). We have previously shown that inducing additional ER stress in growth plate chondrocytes, by engineering the Col2a1 promoter–driven expression of misfolded and intracellularly retained thyroglobulin, is enough to induce a decrease in chondrocyte proliferation and resultant dwarfism independent of apoptosis (Rajpar et al. 2009; Gualeni et al. 2013). Here, we show that disrupting the chondrocyte UPR machinery, by removing a single UPR component, induces ER stress and has a comparable effect on endochondral ossification.
As previously stated, Manf was the highest upregulated gene detected in the chondrocytes of a mouse model of MED resulting from the aggregation of misfolded mutant matrilin-3 in the ER lumen (Leighton et al. 2007; Nundlall et al. 2010). To determine whether the upregulation of Manf in these ‘MED chondrocytes’ was beneficial or in itself a detrimental side effect of ER dysregulation, we crossed the Manffl/fl Col2Cre+ mice with our Matn3V194D model of MED. Interestingly, the deletion of Manf from Matn3 mutant chondrocytes resulted in an aggravation of the MED skeletal phenotype with the ‘double mutant’ mice exhibiting further shortened bones and bell-shaped rib cages that impeded their breathing. Based on this finding, we postulate that the upregulation of Manf expression in the MED chondrocytes has a beneficial role and may be a potential therapeutic target.
MANF can also be upregulated during tunicamycin-induced ER stress (Liu et al. 2016; Wang et al. 2015) and is secreted under ER stress due to being outcompeted for binding to the intracellular KDEL receptors (Oh-Hashi et al. 2012; Henderson et al. 2013). Exogenous MANF has been shown to have an anti-apoptotic, pro-proliferative and anti-ER stress influence in several cell types (Cunha et al. 2017; Airavaara et al. 2009; Gao et al. 2017). Here, we show a similar effect of exogenous MANF on ER-stressed chondrocytes, as evidenced by the reduced levels of GRP78 and increased cell proliferation of tunicamycin-treated primary chondrocytes following the addition of MANF. However, although the treatment of matrilin-3 mutant cells with exogenous MANF reduced the levels of GRP78, it did not reduce the levels of mutant matrilin-3 retention, indicating that it is the intracellular MANF that plays a beneficial role in the pathobiology of matrilin-3 related MED.
To conclude, these data, in conjunction with our previous studies, demonstrates that ER homeostasis is essential for cartilage development and bone growth and that ER stress provides a defined genetically tractable target for therapy.
References
Airavaara M, Shen H, Kuo C-C, Peränen J, Saarma M, Hoffer B, Wang Y (2009) Mesencephalic astrocyte-derived neurotrophic factor (MANF) reduces ischemic brain injury and promotes behavioral recovery in rats. J Comp Neurol 515:116–124
Apostolou A, Shen Y, Liang Y, Luo J, Fang S (2008) Armet, a UPR-upregulated protein, inhibits cell proliferation and ER stress-induced cell death. Exp Cell Res 314:2454–2467
Balasubramanian MN, Butterworth EA, Kilberg MS (2013) Asparagine synthetase: regulation by cell stress and involvement in tumor biology. Am J Physiol Endocrinol Metab 304:E789–E799
Cameron TL, Bell KM, Tatarczuch L, Mackie EJ, Rajpar MH, McDermott BT, Boot-Handford RP, Bateman JF (2011) Transcriptional profiling of chondrodysplasia growth plate cartilage reveals adaptive ER-stress networks that allow survival but disrupt hypertrophy. PLoS One 6:e24600
Cotterill SL, Jackson GC, Leighton MP, Wagener R, Makitie O, Cole W, Briggs MD (2005) Multiple epiphyseal dysplasia mutations in MATN3 cause misfolding of the A-domain and prevent secretion of mutant matrilin-3. Hum Mutat 26:557–567
Craig R, Cortens JP, Beavis RC (2004) Open source system for analyzing, validating, and storing protein identification data. J Proteome Res 3:1234–1242
Crawford RR, Prescott ET, Sylvester CF, Higdon AN, Shan J, Kilberg MS, Mungrue IN (2015) Human CHAC1 protein degrades glutathione, and mRNA induction is regulated by the transcription factors ATF4 and ATF3 and a bipartite ATF/CRE regulatory element. J Biol Chem 290:15878–15891
Cunha DA, Cito M, Grieco FA, Cosentino C, Danilova T, Ladriere L, Lindahl M, Domanskyi A, Bugliani M, Marchetti P, Eizirik DL, Cnop M (2017) Pancreatic beta-cell protection from inflammatory stress by the endoplasmic reticulum proteins thrombospondin 1 and mesencephalic astrocyte-derived neutrotrophic factor (MANF). J Biol Chem 292:14977–14988
Dixon J, Dixon MJ (2004) Genetic background has a major effect on the penetrance and severity of craniofacial defects in mice heterozygous for the gene encoding the nucleolar protein treacle. Dev Dyn 229:907–914
Fang N, Zhang W, Xu S, Lin H, Wang Z, Liu H, Fang Q, Li C, Peng L, Lou J (2014) TRIB3 alters endoplasmic reticulum stress-induced beta-cell apoptosis via the NF-kappaB pathway. Metabolism 63:822–830
Fels DR, Koumenis C (2006) The PERK/eIF2alpha/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol Ther 5:723–728
Flodby P, Li C, Liu Y, Wang H, Marconett CN, Laird-Offringa IA, Minoo P, Lee AS, Zhou B (2016) The 78-kD glucose-regulated protein regulates endoplasmic reticulum homeostasis and distal epithelial cell survival during lung development. Am J Respir Cell Mol Biol 55:135–149
Fujimori T, Suno R, Iemura S-i, Natsume T, Wada I, Hosokawa N (2017) Endoplasmic reticulum proteins SDF2 and SDF2L1 act as components of the BiP chaperone cycle to prevent protein aggregation. Genes Cells 22:684–698
Fukuda S, Sumii M, Masuda Y, Takahashi M, Koike N, Teishima J, Yasumoto H, Itamoto T, Asahara T, Dohi K, Kamiya K (2001) Murine and human SDF2L1 is an endoplasmic reticulum stress-inducible gene and encodes a new member of the Pmt/rt protein family. Biochem Biophys Res Commun 280:407–414
Gál I, Varga T, Szilágyi I, Balázs M, Schlammadinger J, Szabó G Jr (2000) Protease-elicited TUNEL positivity of non-apoptotic fixed cells. J Histochem Cytochem 48:963–970
Gao FJ, Wu JH, Li TT, Du SS, Wu Q (2017) Identification of mesencephalic astrocyte-derived neurotrophic factor as a novel neuroprotective factor for retinal ganglion cells. Front Mol Neurosci 10:76
Gjymishka A, Su N, Kilberg MS (2009) Transcriptional induction of the human asparagine synthetase gene during the unfolded protein response does not require the ATF6 and IRE1/XBP1 arms of the pathway. Biochem J 417:695–703
Gualeni B, Rajpar MH, Kellogg A, Bell PA, Arvan P, Boot-Handford RP, Briggs MD (2013) A novel transgenic mouse model of growth plate dysplasia reveals that decreased chondrocyte proliferation due to chronic ER stress is a key factor in reduced bone growth. Dis Model Mech 6:1414–1425
Hartley CL, Edwards S, Mullan L, Bell PA, Fresquet M, Boot-Handford RP, Briggs MD (2013) Armet/Manf and Creld2 are components of a specialized ER stress response provoked by inappropriate formation of disulphide bonds: implications for genetic skeletal diseases. Hum Mol Genet 22:5262–5275
Hellman M, Arumae U, Yu LY, Lindholm P, Peranen J, Saarma M, Permi P (2011) Mesencephalic astrocyte-derived neurotrophic factor (MANF) has a unique mechanism to rescue apoptotic neurons. J Biol Chem 286:2675–2680
Henderson MJ, Richie CT, Airavaara M, Wang Y, Harvey BK (2013) Mesencephalic astrocyte-derived neurotrophic factor (MANF) secretion and cell surface binding are modulated by KDEL receptors. J Biol Chem 288:4209–4225
Hino K, Saito A, Kido M, Kanemoto S, Asada R, Takai T, Cui M, Cui X, Imaizumi K (2014) Master regulator for chondrogenesis, Sox9, regulates transcriptional activation of the endoplasmic reticulum stress transducer BBF2H7/CREB3L2 in chondrocytes. J Biol Chem 289:13810–13820
Huang J, Chen C, Gu H, Li C, Fu X, Jiang M, Sun H, Xu J, Fang J, Jin L (2016) Mesencephalic astrocyte-derived neurotrophic factor reduces cell apoptosis via upregulating GRP78 in SH-SY5Y cells. Cell Biol Int 40:803–811
Hughes A, Oxford AE, Tawara K, Jorcyk CL, Oxford JT (2017) Endoplasmic reticulum stress and unfolded protein response in cartilage pathophysiology; contributing factors to apoptosis and osteoarthritis. Int J Mol Sci 18
Kang X, Yang W, Feng D, Jin X, Ma Z, Qian Z, Xie T, Li H, Liu J, Wang R, Li F, Li D, Sun H, Wu S (2017) Cartilage-specific autophagy deficiency promotes ER stress and impairs chondrogenesis in PERK-ATF4-CHOP-dependent manner. J Bone Miner Res 32:2128–2141
Kessner D, Chambers M, Burke R, Agus D, Mallick P (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24:2534–2536
Leighton MP, Nundlall S, Starborg T, Meadows RS, Suleman F, Knowles L, Wagener R, Thornton DJ, Kadler KE, Boot-Handford RP, Briggs MD (2007) Decreased chondrocyte proliferation and dysregulated apoptosis in the cartilage growth plate are key features of a murine model of epiphyseal dysplasia caused by a matn3 mutation. Hum Mol Genet 16:1728–1741
Lindahl M, Danilova T, Palm E, Lindholm P, Voikar V, Hakonen E, Ustinov J, Andressoo JO, Harvey BK, Otonkoski T, Rossi J, Saarma M (2014) MANF is indispensable for the proliferation and survival of pancreatic beta cells. Cell Rep 7:366–375
Liu CL, Zhong W, He YY, Li X, Li S, He KL (2016) Genome-wide analysis of tunicamycin-induced endoplasmic reticulum stress response and the protective effect of endoplasmic reticulum inhibitors in neonatal rat cardiomyocytes. Mol Cell Biochem 413:57–67
McLaughlin M, Karim SA, Montague P, Barrie JA, Kirkham D, Griffiths IR, Edgar JM (2007) Genetic background influences UPR but not PLP processing in the rumpshaker model of PMD/SPG2. Neurochem Res 32:167–176
Meunier L, Usherwood YK, Chung KT, Hendershot LM (2002) A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell 13:4456–4469
Mizobuchi N, Hoseki J, Kubota H, Toyokuni S, Nozaki J, Naitoh M, Koizumi A, Nagata K (2007) ARMET is a soluble ER protein induced by the unfolded protein response via ERSE-II element. Cell Struct Funct 32:41–50
Mungrue IN, Pagnon J, Kohannim O, Gargalovic PS, Lusis AJ (2009) CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol 182:466–476
Nundlall S, Rajpar MH, Bell PA, Clowes C, Zeeff LA, Gardner B, Thornton DJ, Boot-Handford RP, Briggs MD (2010) An unfolded protein response is the initial cellular response to the expression of mutant matrilin-3 in a mouse model of multiple epiphyseal dysplasia. Cell Stress Chaperones 15:835–849
Oh-Hashi K, Tanaka K, Koga H, Hirata Y, Kiuchi K (2012) Intracellular trafficking and secretion of mouse mesencephalic astrocyte-derived neurotrophic factor. Mol Cell Biochem 363:35–41
Ozawa K, Kuwabara K, Tamatani M, Takatsuji K, Tsukamoto Y, Kaneda S, Yanagi H, Stern DM, Eguchi Y, Tsujimoto Y, Ogawa S, Tohyama M (1999) 150-kDa oxygen-regulated protein (ORP150) suppresses hypoxia-induced apoptotic cell death. J Biol Chem 274:6397–6404
Petrova P, Raibekas A, Pevsner J, Vigo N, Anafi M, Moore MK, Peaire AE, Shridhar V, Smith DI, Kelly J, Durocher Y, Commissiong JW (2003) MANF: a new mesencephalic, astrocyte-derived neurotrophic factor with selectivity for dopaminergic neurons. J Mol Neurosci 20:173–188
Piróg KA, Irman A, Young S, Halai P, Bell PA, Boot-Handford RP, Briggs MD (2014) Abnormal chondrocyte apoptosis in the cartilage growth plate is influenced by genetic background and deletion of CHOP in a targeted mouse model of pseudoachondroplasia. PLoS One 9:e85145
Pirog-Garcia KA, Meadows RS, Knowles L, Heinegard D, Thornton DJ, Kadler KE, Boot-Handford RP, Briggs MD (2007) Reduced cell proliferation and increased apoptosis are significant pathological mechanisms in a murine model of mild pseudoachondroplasia resulting from a mutation in the C-terminal domain of COMP. Hum Mol Genet 16:2072–2088
Qian B, Wang H, Men X, Zhang W, Cai H, Xu S, Xu Y, Ye L, Wollheim CB, Lou J (2008) TRIB3 [corrected] is implicated in glucotoxicity- and endoplasmic reticulum-stress-induced [corrected] beta-cell apoptosis. J Endocrinol 199:407–416
Rajpar MH, McDermott B, Kung L, Eardley R, Knowles L, Heeran M, Thornton DJ, Wilson R, Bateman JF, Poulsom R, Arvan P, Kadler KE, Briggs MD, Boot-Handford RP (2009) Targeted induction of endoplasmic reticulum stress induces cartilage pathology. PLoS Genet 5:e1000691
Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW (2017) ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18:529
Sakai K, Hiripi L, Glumoff V, Brandau O, Eerola R, Vuorio E, Bosze Z, Fassler R, Aszodi A (2001) Stage-and tissue-specific expression of a Col2a1-Cre fusion gene in transgenic mice. Matrix Biol 19:761–767
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682
Shridhar R, Shridhar V, Rivard S, Siegfried JM, Pietraszkiewicz H, Ensley J, Pauley R, Grignon D, Sakr W, Miller OJ, Smith DI (1996a) Mutations in the arginine-rich protein gene, in lung, breast, and prostate cancers, and in squamous cell carcinoma of the head and neck. Cancer Res 56:5576–5578
Shridhar V, Rivard S, Shridhar R, Mullins C, Bostick L, Sakr W, Grignon D, Miller OJ, Smith DI (1996b) A gene from human chromosomal band 3p21.1 encodes a highly conserved arginine-rich protein and is mutated in renal cell carcinomas. Oncogene 12:1931–1939
Shridhar V, Rivard S, Wang X, Shridhar R, Paisley C, Mullins C, Beirnat L, Dugan M, Sarkar F, Miller OJ, Vaitkevicius VK, Smith DI (1997) Mutations in the arginine-rich protein gene (ARP) in pancreatic cancer. Oncogene 14:2213–2216
Suleman F, Gualeni B, Gregson HJ, Leighton MP, Piróg KA, Edwards S, Holden P, Boot-Handford RP, Briggs MD (2011) A novel form of chondrocyte stress is triggered by a COMP mutation causing pseudoachondroplasia. Hum Mutat 33:218–231
Tadimalla A, Belmont PJ, Thuerauf DJ, Glassy MS, Martindale JJ, Gude N, Sussman MA, Glembotski CC (2008) Mesencephalic astrocyte-derived neurotrophic factor is an ischemia-inducible secreted endoplasmic reticulum stress response protein in the heart. Circ Res 103:1249–1258
Tanaka H, Shimada Y, Harada H, Shinoda M, Hatooka S, Imamura M, Ishizaki K (2000) Polymorphic variation of the ARP gene on 3p21 in Japanese esophageal cancer patients. Oncol Rep 7:591–593
Tiwari A, Schuiki I, Zhang L, Allister EM, Wheeler MB, Volchuk A (2013) SDF2L1 interacts with the ER-associated degradation machinery and retards the degradation of mutant proinsulin in pancreatic beta-cells. J Cell Sci 126:1962–1968
Wang H, Wang X, Ke ZJ, Comer AL, Xu M, Frank JA, Zhang Z, Shi X, Luo J (2015) Tunicamycin-induced unfolded protein response in the developing mouse brain. Toxicol Appl Pharmacol 283:157–167
Wu YB, Li HQ, Ren MS, Li WT, Lv XY, Wang L (2013) CHOP/ORP150 ratio in endoplasmic reticulum stress: a new mechanism for diabetic peripheral neuropathy. Cell Physiol Biochem 32:367–379
Xiong Z, Jiang R, Li X, Liu Y, Guo F (2015) Different roles of GRP78 on cell proliferation and apoptosis in cartilage development. Int J Mol Sci 16:21153–21176
Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Caverner DR (2002) The PERK eukaryotic initiation factor 2α kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol 22:3864–3874
Zhang C, Hong FF, Wang CC, Li L, Chen JL, Liu F, Quan RF, Wang JF (2017) TRIB3 inhibits proliferation and promotes osteogenesis in hBMSCs by regulating the ERK1/2 signaling pathway. Sci Rep 7:10342
Zhao H, Liu Y, Cheng L, Liu B, Zhang W, Guo YJ, Nie L (2013) Mesencephalic astrocyte-derived neurotrophic factor inhibits oxygen-glucose deprivation-induced cell damage and inflammation by suppressing endoplasmic reticulum stress in rat primary astrocytes. J Mol Neurosci 51:671–678
Funding
This work was funded by grants from the Wellcome Trust (Grant 084353/Z/07/) and EU (SYBIL—European Community’s Seventh Framework Programme (FP7/2007–2013) under grant agreement no. 602300).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All experiments were performed in compliance with the Scientific Procedures Act of 1986 and the relevant Home Office (under PPL 40/2884 and PPL60/04525) and Institutional regulations governing animal breeding and handling.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Bell, P.A., Dennis, E.P., Hartley, C.L. et al. Mesencephalic astrocyte-derived neurotropic factor is an important factor in chondrocyte ER homeostasis. Cell Stress and Chaperones 24, 159–173 (2019). https://doi.org/10.1007/s12192-018-0953-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-018-0953-7