Abstract
In the past few decades, genome-based approaches have contributed significantly to vaccine development. Our aim was to identify the most conserved and immunogenic antigens of Streptococcus pneumoniae, which can be potential vaccine candidates in the future. BLASTn was done to identify the most conserved antigens. PSORTb 3.0.2 was run to predict the subcellular localization of the proteins. B cell epitope prediction was done for the immunogenicity testing. Finally, BLASTp was done for verifying the extent of similarity to human proteome to exclude the possibility of autoimmunity. Proteins failing to comply with the set parameters were filtered at each step. Based on the above criteria, out of the initial 22 pneumococcal proteins selected for screening, pavB and pullulanase were the most promising candidate proteins.
Similar content being viewed by others
Introduction
Streptococcus pneumoniae (pneumococcus) is a bacterial pathogen that affects children and adults worldwide [1]. It causes many types of invasive diseases such as pneumonia and meningitis especially among children, elders, and immune-deficient patients. Resistance of S. pneumoniae to a variety of antibacterial agents including quinolones, penicillins, macrolides, and cephalosporins has been reported by a recent study [2]. Currently, up to 40 % of clinical infections are caused by a pneumococcal strain resistant to at least one drug, and 15 % are due to a strain resistant to three or more drugs [3]. Pneumococcal diseases are ‘vaccine preventable diseases,’ and the preventative strategies available include targeted use of the 23-valent polysaccharide pneumococcal vaccine for individuals above the age of 2 years. Regular immunization of newborns and children with the 7-valent polysaccharide-protein conjugate pneumococcal vaccine is another way to prevent the disease, though these are poorly efficacious in infants, immunocompromised patients, and the elderly, the populations that are most at risk [1]. The major drawback of a currently available vaccination includes the unaffordable prices and shortage of supply to the countries that are most affected [4].
A polysaccharide capsule completely encompasses S. pneumoniae and plays a major role in its virulence. These virulence factors present in the cell wall attach to tissue in the body and allow the bacteria to survive against immune responses [5]. During invasion, the capsule is an essential determinant of virulence [6]. By preventing complement C3b opsonization of the bacterial cells, the capsule prevents phagocytosis. One hundred five different capsule types of pneumococci have been identified which forms the basis of antigenic serotyping of the organism [7]. Vaccines that act against pneumococcal infections are based on formulations of various capsular (polysaccharide) antigens derived from the highly prevalent serotypes. Thus, none of the current vaccine is effective against all the 105 different serotypes. In addition, other limitations such as serotype replacement [8] by non-vaccine serotypes are reported; hence, an effective protein vaccine against S. pneumoniae is required to prevent pneumococcal infections [7].
Identification and development of vaccines by conventional or traditional method, though successful most times, is a complex process which involves culturing of organisms, isolating an antigen which is specific to that organism, and inactivating and reinjecting it into subjects to check for immune response. Hence, the process is time consuming and cost ineffective with a very low success rate and is difficult for organisms which cannot be cultured in vitro. And it is almost impossible to identify and develop a universal vaccine candidate using a conventional method for variable organisms like S. pneumoniae containing various strains. This would require the tedious process of searching for an antigen that is common to all strains, processing it, and testing it for vaccine potency [9].
With the introduction of genomic technologies such as whole genomic sequencing, recombinant DNA technology, in silico analysis, and proteomics, the way of studying bacterial pathogenesis and vaccine design has been revolutionized [10]. These advances in genomic technologies mean that every single antigen of a pathogen can be tested for its ability to induce a protective immune response. Reverse vaccinology (RV) is one such approach which is reverse to the conventional approach and involves the mining of genome information of the organism using a proteomic and bioinformatics-based software [11]. This overcomes the complications of the conventional vaccine candidate screening. Here, we have used this novel approach for the identification of the most conserved protein antigens of S. pneumoniae which can be potent vaccine candidates.
Materials
Nucleotide BLAST (BLASTn)
BLAST (basic local alignment search tool) is the most widely used pairwise alignment tool to compare the similarity among two or more nucleotide or protein sequences. It is heuristic which starts alignment from short regions which are similar called hot spots. It gives statistical significance of the result as “expect value” [12]. The several standard output formats include several query-anchored multiple sequence alignment formats, a default pairwise alignment, a taxonomically organized output, and an easily parsable hit table. MegaBLAST, which is the default search program for BLASTn page, operates ten times faster than standard BLAST and finds approximately exact matches [13]. It is designed to find significant alignments among sequences from the same species [14].
PSORTb 3.0.2
Based on the subcellular localization of a protein, it is possible to predict its function, and therefore, a computational tool which can predict a protein’s subcellular localization can be of great help in genome analysis. Since cell surface proteins are important vaccine or drug targets in the case of microbial pathogens, predicting these proteins on the bacterial cell surface could be of great use to develop newer targets. The occurrence of a membrane spanning alpha helices or signal peptides within the primary structure of a protein is an important player that influences a protein’s subcellular localization in a cell. Using these key structural features, PSORTb automatically predicts the subcellular localization of a bacterial protein, whether the protein is present within the cell or localized on the surface [11]. A given protein sequence is examined for presence of motifs, signal peptides, and transmembrane alpha helices, and compared to similar proteins whose localization is previously known. Using a probabilistic technique, the above features are analyzed, and a set of five likely localization sites are returned with their respective probability scores. In fivefold cross-validation analyses, a high overall precision (specificity) of 97 % and a relatively high recall (sensitivity) of 75 % were attained, validating the accuracy of PSORTb.
The subcellular localization of ten selected proteins was done by PSORTb 3.0.2 [11] to check whether the proteins were indeed surface expressed or not. To assess whether a protein is expressed at a specific location, a probabilistic study and fivefold cross-validation study are performed to provide the prediction site of a particular module. The probability values of the five localization sites of a query protein are generated using the above likelihoods.
Kolaskar and Tongaonkar Antigenicity Scale
A simple molecule or a macromolecule such as protein or a lipopolysaccharide or whole cell that can be tightly bound to an antibody or a B cell receptor is termed an antigen. In an antigen, the whole molecule does not bind to the receptor or the antibody, instead only a part of it binds and is called a B cell epitope, and thus acts as an antigenic determinant. For an antigenic protein, this epitope may be either a group of atoms on the protein surface or a short peptide both present in the three-dimensional space. In this study, we used the Kolaskar and Tongaonkar approach from the Immune Epitope Database (IEDB) where B cell antibody epitopes are predicted using amino acid propensities [15]. On the basis of physiochemical parameters of amino acid residues and their occurring frequencies in known investigational epitopes, this semi-empirical approach was developed and could predict the B cell epitopes with about 75 % accuracy. The most important consideration from this antigenicity scale is that the amino acids Cys, Val, and Leu have high antigenic propensity (Ap) values and are hydrophobic. Hence, whenever these occur on surface, they are most likely to be a part of antigenic determinant.
Protein BLAST (BLASTp)
The potential antigenic proteins short listed above must have no significant similarity to the human proteome to avoid any autoimmunity issues which can be a hurdle in the vaccine development. The bacterial proteins used as vaccine candidates may elicit an immune reaction if they have much similarity to any of the human proteins. BLASTp was performed to confirm the foreignness property of the selected proteins. This sequence similarity tool, which compares the query sequence to the plethora of sequences available in the databases, is available as a web-based tool (http://blast.ncbi.nlm.nih.gov/). It is heuristic which starts the alignment from the short matches among sequences termed as hot spots [12].
Methodology
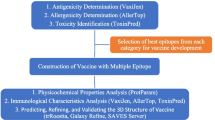
Taking into account the basic principles of RV, the following sequence of steps were applied to screen the vaccine candidates:
Work Flow

Sequence Retrieval
After extensive literature and database search regarding cell surface and antigenic proteins of S. pneumoniae, 22 proteins, covering all classes of surface proteins, were selected for study. The coding DNA sequences of some selected cell surface proteins with antigenic properties were retrieved from GenBank.
Sequence Alignment with Streptococcus pneumoniae Strains
Twenty two proteins [cbpD, codY, hyaluronidase, lytB, lytC, IgA1 protease, pavB, pullulanase (spuA), stkP, nanA, nanB, phpA, phtA, phtE, zmpB, zmpC, FBA, 6GPD, papA, piaA, lytA, and zmpD] were initially selected for doing BLASTn. Accession numbers for the coding DNA sequence (CDS) of the above genes are given in Table 1. BLASTn was run for doing alignment of gene sequences of these proteins. Alignment was restricted to S. pneumoniae (taxid 1313) from the organism search set and “maximum target sequences” were restricted to 50 from the algorithm parameters to provide the most significant results. The cutoff for the “query coverage” percentage to be considered as significant was taken to be 98 %. The BLAST result for every protein was compared for selecting the sequences showing greatest “maximum identity” percentages among maximum number of strains of S. pneumoniae covered.
Subcellular Localization Prediction
CDS for the proteins were translated using the translate tool in the ExPASy resource portal (http://web.expasy.org/translate/). Protein sequences for some of the proteins viz. FBA (P0A4S2) and lytC (Q8DP07) were retrieved from UniProtKB (http://www.uniprot.org/). These protein sequences are then analyzed for their functional localization in the cell using PSORTb software. A cutoff of 7.5 was considered to be good above which a single localization can be assigned. Precision and recall values for the program are calculated using this cutoff.
PSORTb 3.0.2 was run choosing “bacteria” as the “organism” and “Gram positive” was chosen in the “Gram stain” option.
B Cell Epitope Prediction
The “average antigenic score” for all the predicted peptides and the “maximum antigenic score” for the peptides were considered as the selection criteria for the best antigenic proteins. The former value shows the average of all the predicted peptides using a default “window size” of peptides (Default—7), while the latter shows the maximum score from all the peptide segments predicted. The threshold antigenic score is considered to be 1.000. If the average for the whole protein is above 1.0, then the concerned protein is considered to be potentially antigenic. Proteins having relatively greater score than the threshold limit were selected from the initial analysis of five antigenic proteins.
Pairwise Alignment with Human Proteome
The alignment was restricted to the Homo sapiens (taxid 9606) from the organism search set category and “maximum target sequences” were restricted to 50 from the algorithm parameters. In BLASTp, the proteins with no significant similarity (less than 35 %) were considered to be sufficiently distant to the human proteome and will not elicit the antibodies against the self proteins when used as a vaccine candidate [16].
Results
Sequence Alignment with Streptococcus pneumoniae Strains
The gene sequences showing the greatest maximum identity values were selected from these 22 gene sequences. Incorporating the above parameters, the program was run whereby ten proteins viz. cbpD, codY, FBA, lytC, nanB, pavB, phpA, phtE, spuA, and stkP were selected. Alignment results from the BLASTn program for the selected proteins are shown in Table 2.
Prediction of Subcellular Localization of Proteins by PSORTb 3.0.2
Eight out of the ten selected proteins were found to be surface localized with a score higher than 7.5. Out of these, codY, FBA, and stkP were predicted to be cytoplasmic in localization. For lytC, phpA, and phtE, localization scores for cytoplasmic membrane, cell wall, and extracellular region were found to be same, i.e., 3.33; therefore, final prediction for these proteins was given as ‘unknown’ in PSORTb. Two proteins were found to be surface localized [pavB and pullulanase (spuA)] and two proteins (cbpD and nanB) were found to be extracellularly localized. These proteins were selected which were then taken to the next step of the computational analysis. The results of the PSORTb analysis are given in Table 3.
Prediction of B Cell Epitopes by Immune Epitope Database (IEDB)
Total number of predicted peptides from cbpD, nanB, pavB, and spuA was 13, 31, 49, and 49, respectively. The average antigenic score for these proteins from the predicted peptides were predicted to be 1.013, 1.015, 1.010, and 1.006, respectively. Highest value for the maximum antigenic score was found to be for cbpD. It also gave the sequence of the peptide fragments for a given protein having the maximum antigenic score.
Based on this analysis, all four proteins, pavB, spuA (pullulanase), nanB, and cbpD, were found to be having considerably higher score than the threshold value. Tables 4 and 5 show the results of the antigenicity property of the analyzed proteins.
Pairwise Alignment with Human Proteome Using BLASTp Program
From the alignment results, two surface proteins, pavB and pullulanase (spuA), were selected. Alignment results are shown in i and ii in Fig. 1 and in i and ii in Fig. 2.
i BLASTp results of choline-binding protein D (cbpD). BLAST hits on query sequence shows alignments with the human proteome. Maximum identity for each human protein sequence with the query sequence is shown in the BLAST table. ii BLASTp results showing significant alignments of the neuraminidase B (nanB). BLAST hits on query sequence shows alignments with the human proteome. Maximum identity for each human protein sequence with the query sequence is shown in the BLAST table
i BLASTp results showing significant alignments of the pneumococcal adherence and virulence factor (pavB) [all results shown]. BLAST hits on query sequence shows alignments with the human proteome. Maximum identity for each human protein sequence with the query sequence is shown in the BLAST table. ii BLASTp results showing significant alignments of the pullulanase (spuA) [all results shown]. BLAST hits on query sequence shows alignments with the human proteome. Maximum identity for each human protein sequence with the query sequence is shown in the BLAST table
Discussion
S. pneumoniae is a variable organism with numerous serotypes constantly being discovered worldwide. A study pertaining to identification of a vaccine candidate for a wide range of S. pnuemoniae strains has not yet been discovered. The novelty of the present study lies in the exploration of genomics in identifying the common vaccine candidate for a range of S. pnuemoniae strains instead of an individual organism. This is termed “pan genome reverse vaccinology” where various strains of an organism are taken into consideration while developing a vaccine candidate [17]. This approach has been used by Maione et al. (2005) where eight strains of group B streptococcus were cloned and tested in the search for highly efficacious vaccine candidates [18]. An approach combining wet lab and dry lab for determining potential vaccine candidates against S. pneumoniae using a genome of a single strain was used to develop new vaccine candidates against the organism [19]. The first classical example of application of reverse vaccinology to identify a potential vaccine candidate was published in the year 2000 involving serogroup B meningococcus (Neisseria meningitides) [20]. Later, a universal vaccine for serogroup B meningococcus was identified during 2006 and in 2011 clinical trials for multicomponent (4CMenB) and recombinant vaccine testing (rMenB) in human beings was undertaken with promising results [21, 22].
Besides bacteria, this approach has been tested against a variety of pathogenic organisms. Kholia et al. demonstrated the usefulness of this approach against protozoan Leishmania through proteome screening by applying useful parameters such as subcellular localization, non-homology to human proteome, and binding affinity with MHC [23]. In another approach, known antigenic spike protein of the SARS coronavirus was selected, and its sequence is analyzed for the areas having maximum antigenicity, which were then optimized via energy minimization for better antigenic property [16]. Molecular modeling studies by glycoconjugate addition on 14 epitopes of Schistosoma mansoni showed 11 thermodynamically stable epitopes equivalent to native proteins which can be taken for wet lab experiments as vaccine targets against schistosomiasis [24]. A web-based strategy for predicting potential candidates in ten surface-exposed adhesion proteins of Staphylococcus aureus causing endocarditis involved searching for a common nonamer epitope eliciting both humoral and cell-mediated immunity as well as conserved across the various bacterial strains [25].
With the sequences of individual surface proteins and whole genome sequences of pathogenic organisms constantly being discovered, simple application of software is sufficient to screen a particular antigen or a set of antigens within a short span. RV is also cost effective, where genomic data and most software required for screening are easily accessible. Moreover, RV can be applied for a wide range of bacterial, fungal, viral, and parasitic pathogens. This approach uses the BLAST tools which can be performed universally for every organism ranging from virus to fungi. The IEDB B cell epitope prediction tool analyzes input sequences without any bias for the type of organism. Protein localization tools such as Virus-PLoc [26] and Virus-mPLoc [27] are available for subcellular locality prediction of viral proteins. Eukaryotic protein prediction tools viz. SCLpred [28] and PA-SUB [29] can be used for determining sites of fungal proteins.
Though reverse vaccinology has many advantages, it has its own share of disadvantages. Primarily, whole genomic information related to the pathogen should be available, and most importantly, identified antigens are to be backed with in vivo tests to validate the vaccine candidates.
The main aim of the study was to exploit the web-based bioinformatics tools to predict the most conserved and potential antigenic proteins of S. pneumoniae. Various studies have been done previously on a number of pneumococcal surface proteins for their antigenic potential and immunogenicity in animal models. Proteins like pspA and pspC have elicited protective immunity in mice [30]. Recombinant pneumococcal glycolytic enzymes such as FBA and GAPDH (Glyceraldehyde 3-phosphate dehydrogenase) expressed in Escherichia coli [31], produce antibodies that react with various serotypes. Antibodies against CBPs (choline-binding proteins) interact with pneumococcal serotypes as reported by Gosink et al. [32]. Immunization with spuA and phpA [33] generate antibodies against pneumococci.
Four significant properties of any antigenic protein were considered—conservedness, immunogenicity, surface localization, and unrelatedness with human proteome. For evaluating the conservedness of the selected proteins across the pneumococcal serotypes, we have used BLASTn program for the selection of proteins showing maximum identity among the sequences for all strains available in the database. Surface-located proteins have been of much use as vaccine candidates for many years due to their great immunogenic capability. So, we have predicted the surface localization through PSORTb 3.0.2. Immunogenic potential of the candidate proteins were calculated via the ‘Kolaskar and Tongaonkar antigenicity scale’ in the IEDB which gives an antigenic score for different peptides in a protein. BLASTp program in the National Centre for Biotechnology Information (NCBI) was used for the sequence alignment of the proteins with the sequences of the available human proteins in the databases. The notable advantage of the computational analysis for the antigen prediction is the screening of numerous candidates for the selection of most promising among them. This approach can serve as a first-pass filter in the process of vaccine development. This analysis gave most convincing results for the two surface proteins, pavB and spuA. pavB is encoded by open reading frame SP0082 and derived its name from another fibronectin binding protein termed as pavA [34]. A major factor which determines virulence in Gram-positive cocci is their ability to interact with serum or ECM (extracellular matrix) proteins. TIGR strain genome annotation showed four streptococcal surface repeats (SSUREs) in pavB, whereby the third SSURE binds to immobilized fibronectin, and the separate SSUREs bind with repetitive motifs of a host’s fibronectin [34, 35]. pavB has previously been identified as an important bacterial surface protein that binds to fibronectin and plasminogen components of the extracellular matrix and, therefore, plays a central role in molecular pathogenesis of pneumococcal infections [36]. Studies like these have established pavB as a vital protein component of S. pneumoniae. Studies by Jensch et al. (2010) demonstrated pavB as a surface-exposed molecule participating in pneumococcal colonization, where pavB works as an adhesin. This study demonstrated the presence of pavB in all of the strains under test. A delay in migration to the lungs and decreased colonization rates was observed with mutant pavB strains [37]. Surface protein pullulanase was so named as it was homologous to the 3′ region of gene encoding alkyl amylopullulanase (apuA) in Bacillus sp. [38]. Bongaerts et al. characterized pullulanase (spuA) and found it as highly conserved and is expressed in all growth phases of S. pneumoniae, making it as a potential vaccine candidate in the future [39]. Southern and Western blot analysis identified spuA as monocistronic with a molecular weight of 140 kDa. Furthermore, YNWGY motif, common to all pullulanases, was identified in spuA [39, 40]. Four regions (I, II, III, and IV) along with their catalytic triad, Asp785–Glus814–Asp902, which forms a catalytic domain of amylolytic enzymes, was reported in spuA. Moreover, amino acid sequence revealed presence of LPXTG motif, and surface localization was confirmed via immunocytometry and immunofluorescence. N-terminal region of spuA was found to be located on the surface, capable of eliciting immunity and species specific. Author has emphasized the need of inactivation and virulence studies in deciphering the role of these proteins in the pathogenesis of S. pneumoniae [39]. Structure-function studies of spuA and pulA through X-ray crystallography deciphered the molecular basis of glycogen specificity. This study observed the specificity of these proteins to glycogen of type II alveolar cells and probability of alpha-glucan metabolizing mechanism being a crucial factor responsible for streptococcal virulence [41]. Abbott et al. also characterized two proteins, spuA and MalX, and deduced glycogen as the substrate for these enzymes [42]. These studies suggest that spuA helps in binding to epithelial cells of lung which is a familiar target site for this pathogen. These proteins can be tested further for their immunogenic property in the wet lab experiments for their future use as a well-conserved and highly immunogenic vaccine candidate against a variety of S. pneumoniae strains. The present work, though preliminary, is both cost effective and less time consuming for screening of vaccine candidates when compared to a traditional approach, but the screened vaccine candidate has to be validated using a biological assay.
References
Ogunniyi, A. D., Grabowicz, M., Briles, D. E., Cook, J., & Paton, J. C. (2007). Development of a vaccine against invasive pneumococcal disease based on combinations of virulence proteins of Streptococcus pneumoniae. Infection and Immunity, 75, 350–357.
Jenkins, S. G., Brown, S. D., & Farrell, D. J. (2008). Trends in antibacterial resistance among Streptococcus pneumoniae isolated in the USA. Annals of Clinical Microbiology and Antimicrobials, 7, 1–11.
Eng, J. V., Marcus, R., Hadler, J. L., Imhoff, B., Vugia, D. J., Cieslak, P. R., et al. (2003). Consumer attitudes and use of antibiotics. Emerging Infectious Diseases, 9, 1128–1135.
Ceppa, L.L.P. (2012). Lessons learnt from the Accelerated Vaccine Introduction (AVI) project, GAVI alliance. Available from: http://www.gavialliance.org/results/evaluations/avi-project-review/. Accessed July 7, 2012.
Todar, K. (2003). In Todar’s online textbook of bacteriology: Streptococcus pneumoniae: pneumonia, otitis media and meningitis, available from http://textbookofbacteriology.net/S.pneumoniae.html. Accessed July 9, 2012.
Weinberger, D. M., Malley, R., & Lipsitch, M. (2011). Serotype replacement in disease after pneumococcal vaccination. Lancet, 378, 1962–1973.
Moyo, S. J., Steinbakk, M., Aboud, S., Mkopi, N., Kasubi, M., Blomberg, B., et al. (2012). Penicillin resistance and serotype distribution of Streptococcus pneumoniae in nasopharyngeal carrier children under-5 years in Dar es Salaam, Tanzania. Journal of Medical Microbiology, 61, 952–959.
Jedrzejas, M. J. (2001). Pneumococcal virulence factors: structure and function. Microbiology and Molecular Biology Reviews, 65, 187–207.
Kelly, D. F., & Rappuoli, R. (2004). Reverse vaccinology and vaccines for serogroup B Neisseria meningitides. Advances in Experimental Medicine and Biology, 568, 217–223.
Capecchi, B., Serruto, D., Adu- Bobie, J., Rappuoli, R., & Pizza, M. (2004). The genome revolution in vaccine research. Current Issues in Molecular Biology, 6, 17–28.
Yu, N. Y., Wagner, J. R., Laird, M. R., Melli, G., Rey, S., Lo, R., et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics, 26, 1608–1615.
McGinnis, S., & Thomas, L. M. (2004). BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Research, 32, W20–W25.
Wheeler, D. L., Barrett, T., Benson, D. A., Bryant, S. H., & Canese, K. (2006). Databases resources of the National Center for Biotechnology Information. Nucleic Acids Research, 34, D173–D180.
Sayers, E. W., Barrett, T., Benson, D. A., Bryant, S. H., Canese, K., Chetvernin, V., et al. (2009). Databases resources for the National Center for Biotechnology Information. Nucleic Acids Research, 37, D5–D15.
Kolaskar, A. S., & Tongaonkar, P. C. (1990). A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Letters, 276, 172–174.
Sharma, P., & Bezbaruah, R. L. (2011). Antigenic site detection of the spike protein and designing of a vaccine candidate against SARS coronavirus using reverse vaccinology: an in silico approach. International Journal of Pharmacy and Technology, 3, 3555–3568.
Serruto, D., Serino, L., Masignani, V., & Pizza, M. (2009). Genome-based approaches to develop vaccines against bacterial pathogens. Vaccine, 27, 3245–3250.
Maione, D., Margarit, I., Rinaudo, C. D., Masignani, V., Mora, M., Scarselli, M., et al. (2005). Identification of a universal group B streptococcus vaccine by multiple genome screen. Science, 309, 148–150.
Wizemann, T. M., Heinrichs, J. H., Adamou, J. E., Erwin, A. L., Kunsch, C., Choi, G. H., et al. (2001). Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infection and Immunity, 69, 1593–1598.
Pizza, M., Scarlato, V., Masignani, V., Giuliani, M. M., Aricò, B., Comanducci, M., et al. (2000). Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science, 287, 1816–1820.
Giuliani, M. M., Adu-Bobie, J., Comanducci, M., Arico, B., Savino, S., Santini, L., et al. (2006). A universal vaccine for serogroup B meningococcus. Proceedings of the National Academy of Sciences, 103, 10834–10839.
Toneatto, D., Ismaili, S., Ypma, E., Vienken, K., Oster, P., & Dull, P. (2011). The first use of an investigational multicomponent meningococcal serogroup B vaccine (4CMenB) in humans. Human Vaccines & Immunotherapeutics, 7, 646–653.
John, L., John, G. G., & Kholia, T. (2012). A reverse vaccinology approach for the identification of potential vaccine candidates from Leishmania spp. Applied Biochemistry and Biotechnology, 167, 1340–1350.
de Oliveira Lopes, D., de Oliveira, F. M., do Vale Coelho, I. E., de Oliveira Santana, K. T., Mendonça, F. C., Taranto, A. G., et al. (2013). Identification of a vaccine against schistosomiasis using bioinformatics and molecular modeling tools. Infection, Genetics and Evolution, 20, 83–95. doi:10.1016/j.meegid.2013.08.007.
Oprea, M., & Antohe, F. (2013). Reverse-vaccinology strategy for designing T-cell epitope candidates for Staphylococcus aureus endocarditis vaccine. Biologicals, 41, 148–153. doi:10.1016/j.biologicals.2013.03.001.
Shen, H. B., & Chou, K. C. (2010). Virus-mPLoc: a fusion classifier for viral protein subcellular location prediction by incorporating multiple sites. Journal of Biomolecular Structure and Dynamics, 28, 175–186.
Shen, H. B., & Chou, K. C. (2007). Virus-PLoc: a fusion classifier for predicting the subcellular localization of viral proteins within host and virus-infected cells. Biopolymers, 85, 233–240.
Lu, Z., Szafron, D., Greiner, R., Lu, P., Wishart, D. S., Poulin, B., et al. (2004). Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics, 20, 547–556.
Mooney, C., Wang, Y. H., & Pollastri, G. (2011). SCLpred: protein subcellular localization prediction by N-to-1 neural networks. Bioinformatics, 27, 2812–2819.
Ferreira, D. M., Darrieux, M., Silva, D. A., Leite, L. C., Ferreira, J. M., Jr., Ho, P. L., et al. (2009). Characterization of protective mucosal and systemic immune responses elicited by pneumococcal surface protein pspA & pspC nasal vaccines against a respiratory pneumococcal challenge in mice. Clinical and Vaccine Immunology, 16, 636–645.
Mizrachi, N. Y., Ling, E., Feldman, G., Portnoi, M., Dagan, R., Overweg, K., et al. (2004). Glycolytic enzymes associated with cell surface of Streptococcus pneumoniae are antigenic in humans and elicit protective immune responses in the mouse. Clinical and Experimental Immunology, 138, 290–298.
Gosink, K., Mann, E. R., Guglielmo, C., Tuomanen, E. I., & Masure, H. R. (2000). Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infection and Immunity, 68, 5690–5695.
Zhang, Y., Masi, A. W., Barniak, V., Mountzouros, K., Hostetter, M. K., & Green, B. A. (2001). Recombinant phpA protein, a unique histidine motif-containing protein from Streptococcus pneumoniae, protects mice against intranasal pneumococcal challenge. Infection and Immunity, 69, 3827–3836.
Jedrzejas, M. J. (2007). Unveiling molecular mechanisms of bacterial surface proteins: Streptococcus pneumoniae as a model organism for structural studies. Cellular and Molecular Life Sciences, 64, 2799–2822.
Bumbaca, D., Littlejohn, J. E., Nayakanti, H., Rigden, D. J., Galperin, M. Y., & Jedrzejas, M. J. (2004). Sequence analysis and characterization of a novel fibronectin-binding repeat domain from the surface of Streptococcus pneumoniae. OMICS, 8, 341–356.
Paterson, G. K., & Orihuela, C. J. (2010). Pneumococcal microbial surface components recognizing adhesive matrix molecules targeting of the extracellular matrix. Molecular Microbiology, 77, 1–5.
Jensch, I., Gamez, G., Rothe, M., Ebert, S., Fulde, M., Somplatzki, D., et al. (2010). PavB is a surface-exposed adhesion of Streptococcus pneumoniae contributing to nasopharyngeal colonization and airways infections. Molecular Microbiology, 77, 22–43.
Hatada, Y., Igarashi, K., Ozaki, K., Ara, K., Hitomi, J., Kobayashi, T., et al. (1996). Amino acid sequence and molecular structure of an alkaline amylopullulanase from bacillus that hydrolyzes alpha-1,4 and alpha-1,6 linkages in polysaccharides at different active sites. Journal of Biological Chemistry, 271, 24075–24083.
Bongaerts, R. J., Heinz, H. P., Hadding, U., & Zysk, G. (2000). Antigenicity, expression, and molecular characterization of surface-located pullulanase of Streptococcus pneumoniae. Infection and Immunity, 68, 7141–7143.
Albertson, G. D., McHale, R. H., Gibbs, M. D., & Bergquist, P. L. (1997). Cloning and sequence of a type I pullulanase from an extremely thermophilic anaerobic bacterium, Caldicellulosiruptor saccharolyticus. Biochimica et Biophysica Acta, 1354, 35–39.
van Bueren, A. L., Higgins, M., Wang, D., Burke, R. D., & Boraston, A. B. (2007). Identification and structural basis of binding to host lung glycogen by streptococcal virulence factors. Nature Structural and Molecular Biology, 14, 76–84.
Abbott, D. W., Higgins, M. A., Hyrnuik, S., Pluvinage, B., Lammerts van Bueren, A., & Boraston, A. B. (2010). The molecular basis of glycogen breakdown and transport in Streptococcus pneumoniae. Molecular Microbiology, 77, 183–199.
Acknowledgments
We would like to thank Dr. (Mrs.) Mangala Lahkar for providing necessary resource materials and for timely completion of this work.
Conflicts of Interest Statement
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Sandipan Talukdar, Shubhranshu Zutshi, and K. S. Prashanth contributed equally.
Rights and permissions
About this article
Cite this article
Talukdar, S., Zutshi, S., Prashanth, K.S. et al. Identification of Potential Vaccine Candidates Against Streptococcus pneumoniae by Reverse Vaccinology Approach. Appl Biochem Biotechnol 172, 3026–3041 (2014). https://doi.org/10.1007/s12010-014-0749-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-014-0749-x