Abstract
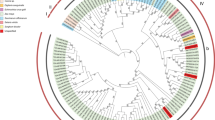
Beet mosaic virus (BtMV), the only Potyvirus known to infect sugar beet, occurs worldwide in beet crops. The full genome sequencing of a BtMV isolate from Iran (Ir-VRU), enabled us to better understand the evolutionary history of this virus. Selection analysis suggested that BtMV evolution is mainly under negative selection but its strength varies in different proteins with the multifunctional proteins under strongest selection. Recombination has played a major role in the evolution of the BtMVs; only the Ir-VRU and USA isolates show no evidence of recombination. The ML phylogenies of BtMVs from coat protein and full sequences were completely congruent. The primary divergence of the BtMV phylogeny is into USA and Eurasian lineages, and the latter then divides to form a cluster only found in Iran, and a sister cluster that includes all the European and Chinese isolates. A simple patristic dating method estimated that the primary divergence of the BtMV population was only 360 (range 260–490) years ago, suggesting an emergence during the development of sugar beet as a crop over the past three centuries rather than with the use of leaf beet as a vegetable for at least 2000 years.



Similar content being viewed by others
References
M.M. Romeiras, A. Vieira, D.N. Silva, M. Moura, A. Santos-Guerra, D. Batista, M.C. Duarte, O.S. Paulo, PLoS ONE (2016). https://doi.org/10.1371/journal.pone.0152456
G.H. Coons, Proc. Am. Soc. Sugar Beet Technol. 8, 2 (1954)
A.S. Marggraf, Histoire De L’académie Royale Des Sciences Et Belles-lettres De Berlin. (Deutsche Akademie der Wissenschaften zu Berlin, 1747), pp. 79–90
P. Hanelt, R. Buttner, R. Mansfeld, Mansfeld’s Encyclopedia of Agricultural and Horticultural Crops (Except Ornamentals) (Springer, Berlin, 2001), pp. 237–240
C. Winner, in The Sugar Beet Crop, ed. By D. A. Cooke, R. K. Scott (Springer, 1993), pp. 1
G.E. Russell, C.M.I./A.A.B. Descr Plant Vir 53, 3 (1971)
R.M. Harveson, L.E. Hanson, G.L. Hein, Compendium of Beet Diseases and Pests, 2nd edn. (APS Press, New York, 2009)
E. Prillieux, G. Delacroix, Acad. Des Sci. Compt. Rend. 127, 6 (1898)
D.D. Sutic, R.E. Ford, M.T. Tosic, Handbook of Plant Virus Diseases (CRC Press, Baco Raton, 1999), pp. 276–291
L.K. Jones, Mosaic Disease of Beets (Washington Agricultural Experiment Station Bulletin, Washington, 1931)
K.M. Smith, Textbook of Plant Virus Diseases, 3rd edn. (Academic Press, Cambridge, 1972), pp. 92–95
S. Veerakone, J.Z. Tang, L.I. Ward, L.W. Liefting, Z. Perez-Egusquiza, B.S.M. Lebas, C. Delmiglio, J.D. Fletcher, P.L. Guy, Australas. Plant. Pathol. (2015). https://doi.org/10.1007/s13313-015-0366-3
A.J. Gibbs, Ann Appl Biol. (1960). https://doi.org/10.1111/j.1744-7348.1960.tb03578.x
A.N. Dusi, D. Peters, J. Phytopathol. (1999). https://doi.org/10.1046/j.1439-0434.1999.147005293.x
H.Y. Wang, X.D. Li, Y.Y. Liu, B. Wang, X.P. Zhu, Plant Pathol. (2008). https://doi.org/10.1111/j.1365-3059.2008.01852.x
R.J. Shepherd, B.B. Till, N. Schaad, J. Am. Sug. Beet Technol. 14, 2 (1966)
S.G. Kumari, A. Najar, N. Attar, M.H. Loh, H.J. Vetten, Plant Dis. (2010). https://doi.org/10.1094/PDIS-94-8-1068C
R. Shepherd, F. Hills, D. Hall, J. Am. Soc. Sug. Beet Technol. 13, 3 (1964)
W.M. Wintermantel, Plant Dis. (2005). https://doi.org/10.1094/PD-89-0325
J. Chen, J. Chen, M.J. Adams, Arch. Virol. (2001). https://doi.org/10.1007/s007050170144
C. Ha, S. Coombs, P.A. Revill, R.M. Harding, M. Vu, J.L. Dale, Arch. Virol. (2008). https://doi.org/10.1007/s00705-007-1053-7
G. Lu, E.N. Moriyama, Brief Bioinform. (2004). https://doi.org/10.1093/bib/5.4.378
J. Ye, G. Coulouris, I. Zaretskaya, I. Cutcutache, S. Rozen, T.L. Madden, BMC Bioinform. (2012). https://doi.org/10.1186/1471-2105-13-134
T.A. Hall, Nucleic Acids Symp. Ser. 41, 41 (1999)
H. Hasan, Generation of an infectious beet mosaic virus (BtMV) full-length clone based on the complete nucleotide sequence of a german isolate (Hannover University, Hannover, 2004)
K. Katoh, D.M. Standley, Mol. Biol. Evol. (2013). https://doi.org/10.1093/molbev/mst010
F. Abascal, R. Zardoya, M.J. Telford, Nucleic Acids Res. (2010). https://doi.org/10.1093/nar/gkq291
D. Darriba, G.L. Taboada, R. Doallo, D. Posada, Nat. Methods (2012). https://doi.org/10.1038/nmeth.2109
D. Darriba, G.L. Taboada, R. Doallo, D. Posada, Bioinformatics (2011). https://doi.org/10.1093/bioinformatics/btr088
A. Stamatakis, Bioinformatics (2014). https://doi.org/10.1093/bioinformatics/btu033
H. Shimodaira, M. Hasegawa, Mol. Biol. Evol. (1999). https://doi.org/10.1093/oxfordjournals.molbev.a026201
G. Yu, D.K. Smith, H. Zhu, Y. Guan, T.T.Y. Lam, Methods Ecol. Evol. (2016). https://doi.org/10.1111/2041-210X.12628
D.P. Martin, B. Murrell, M. Golden, A. Khoosal, Virus Evol. (2015). https://doi.org/10.1093/ve/vev003
S.L. Kosakovsky Pond, S.D. Frost, Mol. Biol. Evol. (2005). https://doi.org/10.1093/molbev/msi105
W. Delport, A.F. Poon, S.D. Frost, S.L.K. Pond, Bioinformatics. (2010). https://doi.org/10.1093/bioinformatics/btq429
A.J. Gibbs, H.D. Nguyen, K. Ohshima, Curr Opin Virol. (2015). https://doi.org/10.1016/j.coviro.2014.12.004
M. Fourment, M.J. Gibbs, BMC Evol. Biol. (2006). https://doi.org/10.1186/1471-2148-6-1
R.J. Jackson, C.U. Hellen, T.V. Pestova, Nat. Rev. Mol. Cell. Biol. (2010). https://doi.org/10.1038/nrm2838
M.J. Adams, J.F. Antoniw, F. Beaudoin, Mol Plant Pathol. (2005). https://doi.org/10.1111/j.1364-3703.2005.00296.x
L.G. Nemchinov, J. Hammond, R. Jordan, R.W. Hammond, Arch. Virol. (2004). https://doi.org/10.1007/s00705-003-0278-3
B.Y.W. Chung, W.A. Miller, J.F. Atkins, A.E. Firth, Proc. Natl. Acad. Sci. (2008). https://doi.org/10.1073/pnas.0800468105
T. Turpen, J. Gen. Virol. (1989). https://doi.org/10.1099/0022-1317-70-8-1951
H. Xiang, Y.H. Han, C. Han, D. Li, J. Yu, Virus Genes (2007). https://doi.org/10.1007/s11262-007-0132-x
A. Gibbs, K. Ohshima, Annu. Rev. Phytopathol. (2010). https://doi.org/10.1146/annurev-phyto-073009-114404
A.J. Gibbs, K. Ohshima, R. Yasaka, M. Mohammadi, M.J. Gibbs, R.A. Jones, Virus Evol. (2017). https://doi.org/10.1093/ve/vex002
H.D. Nguyen, Y. Tomitaka, S.Y. Ho, S. Duchêne, H.J. Vetten, D. Lesemann, J.A. Walsh, A.J. Gibbs, K. Ohshima, PLoS ONE (2013). https://doi.org/10.1371/journal.pone.0055336
B. Desplanque, P. Boudry, K. Broomberg, P. Saumitou-Laprade, J. Cuguen, H. Van Dijk, Theor. Appl. Genet. (1999). https://doi.org/10.1007/s001220051184
S. Fénart, J.F. Arnaud, I. De Cauwer, J. Cuguen, Theor. Appl. Genet. (2008). https://doi.org/10.1007/s00122-008-0735-1
A.A. Brunt, K. Crabtree, M.J. Dallwitz, A.J. Gibbs, L. Watson, Viruses of plants: Descriptions and lists from the VIDE database (Cab International, Wallingford, 1996)
A.J. Gibbs, A.M. Mackenzie, K.J. Wei, M.J. Gibbs, Arch. Virol. (2008). https://doi.org/10.1007/s00705-008-0134-6
M. Arjmand, J. Sugar Beet Res. 30, 4 (1993)
G. Geng, J. Yang, Sugar Tech. (2015). https://doi.org/10.1007/s12355-014-0353-y
Acknowledgements
The authors would like to thank to Tahereh Ramazani for kindly collecting beet samples.
Author information
Authors and Affiliations
Contributions
AH and SH prepared and provided laboratory support, MM amplified and sequenced the IR-VRU isolate, carried out the analyses, and wrote the draft manuscript, AG designed the dating method, interpreted the results, and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Edited by Seung-Kook Choi.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mohammadi, M., Gibbs, A.J., Hosseini, A. et al. An Iranian genomic sequence of Beet mosaic virus provides insights into diversity and evolution of the world population. Virus Genes 54, 272–279 (2018). https://doi.org/10.1007/s11262-018-1533-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-018-1533-8