Abstract
Cathepsins, growth hormone-releasing hormone (GHRH) and leptin receptor (LEPR) genes have been receiving increasing attention as potential markers for meat quality and pig performance traits. This study investigated the allele variants in four cathepsin genes (CTSB, CTSK, CTSL, CTSS), GHRH and LEPR in pure-bred Ukrainian Large White pigs and evaluated effects of the allele variants on meat quality characteristics. The study was conducted on 72 pigs. Genotyping was performed using PCR–RFLP technique. Meat quality characteristics analysed were intramuscular fat content, tenderness, total water content, ultimate pH, crude protein and ashes. A medium level of heterozygosity values was established for GHRH and LEPR genes which corresponded to very high levels of informativeness indexes. Cathepsins CTSL, CTSB and CTSK had a low level of heterozygosity, and CTSS did not segregate in this breed. Association studies established that intramuscular fat content and tenderness were affected by the allele variance in GHRH and LEPR but not by CTSB and CTSL genes. The GHRH results could be particularly relevant for the production of lean prime cuts as the A allele is associated with both, a lower meat fat content and better tenderness values, which are two attributes highly regarded by consumers. Results of this study suggest that selective breeding towards GHRH/AA genotype would be particularly useful for improving meat quality characteristics in the production systems involving lean Large White lines, which typically have less than 2 % intramuscular fat content.
Similar content being viewed by others
Introduction
During the last decades, the international pig industry has been focusing on the selective breeding towards genotypes with high growth rate, improved feed efficiency and increased meat content. Unfortunately, this was accompanied by reduction of intramuscular fat and water holding capacity, which had deteriorating effect on meat eating quality traits such as flavour and juiciness [1]. The industry has been employing a number of strategies to enhance intramuscular fat content and improve meat eating quality characteristics. This includes manipulation of pig diets and revised management procedures during animals’ transportation and slaughter [2, 3]. In parallel, increasing attention has been paid to identifying genetic markers for desirable combination of meat quality and animal performance traits. A number of Quantitative Trait Loci (QTLs) associated with pig meat quality traits and/or carcass composition have been reported (reviewed in [4]) and several cathepsin genes have been identified as potential genetic markers among positional candidate genes. Cathepsins are a family of peptidase enzymes which are present mainly in the lysosomes of the skeletal muscles using actin, myosin and associated proteins as substrates for their reaction [5]. Therefore, cathepsins could play an important role in the post-mortem meat tenderisation [6]. It has been demonstrated that polymorphisms in porcine cathepsin genes are associated with daily gain, backfat thickness, lean content and intramuscular fat content [7–13].
In parallel with cathepsins, the Growth Hormone Releasing Hormone (GHRH), which controls muscle growth and development, has been considered as a candidate gene for pig performance and meat quality traits. Polymorphisms in GHRH gene have been shown to be associated with backfat thickness, daily gain, carcass meat content, meat colour and water-holding capacity [14, 15]. Similarly, mutations in the leptin receptor (LEPR) have been associated with traits directly related to fat deposition, such as backfat thickness, intramuscular fat content, lean content and average daily gain [16–18].
The mechanisms regulating meat quality traits and pig performance are breed-specific due to interaction with genetic background that composes each population [19]. Therefore, it is important to evaluate the strengths of associations between candidate genetic markers and meat quality traits in the breeds and pig lines intended for specific markets. In this regard, Large White pigs are widely used for crossbreeding and to generate new production lines in 117 countries across the world [20, 21]. During the last decades, an increasing attention has been paid to traditional Ukrainian pig breeds and pig lines including Ukrainian Large White. This breed was created in the end of 20th century on the basis of English Large White pig breed using a complex selection process aiming to improve meat quality traits. Ukrainian Large White pigs have multiple pregnancies with an average litter size of 11.6 piglets and a daily weight gain 800–850 g/day. They also have strong body built and are well adapted to difficult climate conditions. Ukrainian Large White pigs have been widely used for production of “organic” pork and outdoor rearing. Depending on the live weight at the slaughter time, Ukrainian Large White pigs can be used for production of a range of products, from lean meat and bacon to traditional Ukrainian “salo”, which is cured slabs of fatback.
In spite of a large number of publications on GHRH and LEPR polymorphisms, there is very limited and fragmental information on associations between there polymorphisms and meat quality and pig performance traits. Previous studies were carried out on common Large White populations from a number of European countries but not on Ukraine Large White pigs [8, 9, 15, 17]. Furthermore, so far no studies have tested associations between cathepsin genes, GHRH and LEPR and meat quality traits such a protein content, tenderness, ashes, and total water in Large White pigs.
The aim of the present study was to investigate the strength of associations between polymorphisms in the cathepsin genes (CTSB, CTSK, CTSL, and CTSS), GHRH and LEPR with meat quality traits in purebred Ukrainian Large White pigs. Specific objectives were: (i) to identify the allele variants of CTSB, CTSK, CTSL, CTSS, GHRH and LEPR in the Ukrainian Large White pigs, and (ii) to evaluate effects of these allele variants on meat quality traits.
Materials and methods
Animals
The study was conducted on 72 pigs of Ukrainian Large White breed. The authors recognise that this number is lower when compared to other association studies. This was due the fact that the size of Ukrainian Large White population is relatively small (around 500 pure bred pigs in Ukraine) and it is not easy to access these animals. Experimental protocol was approved by the Scientific Committee of the Institute of Pig Breeding and Agro-Industrial Production, National Academy of Agricultural Sciences, Ukraine. All the procedure related to animal handling and slaughter were conducted in accordance with the European Convention for the Protection of Vertebrate Animals used for Experimental and Others Scientific Purposes [22].
The study used castrated males or females in approximately 1:1 ratio. The animals were raised at the facilities of the Ukrainian Academy of Agricultural Sciences, Ukraine, fed a standard commercial finishing diet ad libitum from the live weight of 40 kg, were handled by the same staff, and were slaughtered at the average live weight of 109 kg in one season, at the same time of the day and under the same conditions. In the association studies, the gender of animals was present as a random factor. The association studies design was based on the approach described by [23].
During the growth phase between 40 and 60 kg (live weight), the feed contained per dry matter: 12.9 MJ/kg of net energy, 19.1 % of crude protein and 1.14 % of lysine. During the growth phase between 60 and 90 kg (live weight), the feed contained 12.8 MJ/kg of net energy, 18.0 % of crude protein and 1.0 % of lysine. During the finishing phase (live weight 90–109 kg), the feed contained 12.6 MJ/kg of net energy, 17.1 % of crude protein and 0.8 % of lysine. The feed was produced by Poltava Feed Mill (Poltava, Ukraine).
Samples of blood (1 ml) were collected when the pigs reached the weight of 109 kg. The blood was collected in the morning before feeding. The blood samples were mixed with 0.05 M EDTA and stored up to 7 days at +4 °C until used for DNA isolation.
Analysis of meat quality traits
The pigs used in the association studies were tested for the c.1843 C>T mutation in the ryanodine receptor gene associated with pig meat defects [24]. All the animals had genotype CC, e.g. the mutant allele variant was absent.
Ultimate pH was measured by a portable digital LF-Meter “LF-Star CPU-Pistole” (Ing.-Büro & Klassifizierungsservice Rudolf Matthäus, Klausa, Germany) after cooling the carcasses for 48 h at +2 to +4 °C. The analyses were conducted on samples from M. longissimus dorsi collected between the 10th and 12th thoracic vertebra. The samples were collected from the same position on the right side of the carcasses.
The total water content was determined by drying of meat samples at 105 °C to constant weight; ash analysis was carried out by burning the meat samples in a muffle furnace at 550° C; crude protein content was analysed by Kjeldahl’s method, and intramuscular fat content was determent by Soxhlet’s method as described in Official Methods of Analysis [25].
Meat tenderness was determined using the protocol for Warner–Bratzler shear force analysis [26]. For each sample, two cylindrical cores (8 cm long and 2.5 cm in the diameter) were taken in parallel to the longitudinal orientation of the muscle fibres. The cylindrical cores were shredded by a cutting blade at a velocity 200 mm/min at six points. The time of shredding was recorded for each point and averages were calculated to determine the shear force value per sample.
PCR amplification
Genomic DNA was isolated from blood by the sorbent method using Diatom™ DNA Prep 100 kit (Isogen, Russia, Moscow) following the manufacturer’ instructions with guanidine thiocyanate as the lysis reagent. Polymerase Chain Reaction (PCR) for Restriction Fragment Length Polymorphism Analysis of PCR-Amplified Fragments (PCR–RFLP) genotyping was carried out in a final volume of 25 µl which contained 200 nm of corresponding forward and reverse primers, 1.5, 2.0 or 2.5 mM MgCl2, 0.25 mM of each of the dNTP and 1 unit of the recombinant Taq DNA Polymerase (Fermentas, Vilnius, Lithuania).
Genotyping
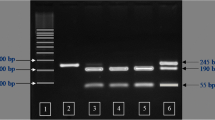
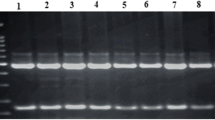
CTSB, CTSL, CTSS and GHRH were genotyped using PCR–RFLP technique as described by [9, 27, 28]. CTSL was genotyped on the g.143T>C SNP (NCBI accession number rs 81212773); CTSS was genotyped on the g.171G>A SNP (NCBI accession number rs 331232519); CTSB was genotyped on the g.72A>C SNP (EMBL accession number AJ315558), and GHRH was genotyped on the AluI polymorphic restriction site in exon 3.
For the CTSL restriction analysis, 5 µl of PCR product was digested overnight at 65 °C with 3 units of TaqI restriction enzyme (Fermentas) in the final volume of 25 µl of the reaction buffer. Digestion of the CTSS PCR product was conducted under conditions similar to that for CTSL but with the BseNI endonuclease (Fermentas). The restriction analysis of CTSB amplification product was carried out by digestion with MspI endonuclease (Fermentas), and the restriction analysis of GHRH amplification product was carried out using the restriction enzyme AluI (Fermentas). The DNA fragments obtained by restrictive digestion, were separated on 4 % agarose gels, and DNA was visualised using ethidium bromide (0.5 µg/µl CTSK g.15G>A SNP (NCBI accession number rs 337183461) and LEPR c.1987C>T SNP (db SNP accession number ss 262803826) were genotyped using allelic discrimination and High Resolution Melting (HRM) methods respectively. For HRM, Luminaris Color HRM qPCR Master Mix (LifeTechnologies, Grand Island, NY, USA) was used. The HRM primers were designed using Primer3Plus program (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) with the qPCR default settings, while the allelic discrimination primers and probes were designed as part of Life Technologies custom assays. Both analyses were conducted in CFX96 Touch™ Real Time PCR Detection System. The primers, probes and PCR conditions are given in Table 1.
Statistical analysis
Allele frequencies, genotype frequencies, polymorphic information content (PIC), and levels of heterozygosity (observed heterozygosity, Ho and expected heterozygosity, He) were calculated with GenAlEx 6.0. software [29]. Analysis of associations between genotypes and meat quality characteristics were conducted by one way ANOVA. Means between groups were compared with a two-tailed t test using JMP12 (SAS Inst. Inc., Cary, NC) and differences were considered significant at P < 0.05.
Additive (A) and Dominance (D) components were calculated using the following equations:
where \(\overline{X}_{11} ,\overline{X}_{12} ,\overline{X}_{22}\) are arithmetic mean value of productivity traits for genotypes 1/1, 1/2 and 2/2.
The effect of alleles 1 and 2 were estimated using the equations \(\begin{aligned} \alpha_{1} = m_{1} - \bar{X} \hfill \\ \alpha_{2} = m_{2} - \bar{X} \hfill \\ \end{aligned}\) where
p and q are allelic frequency 1 and 2 respectively; and \(\overline{X}\) is the total arithmetic mean value for each trait. Allelic substitution effects, \(\frac{\alpha }{2}(1 \to 2)\) were calculated using the equation
Results and discussion
Allele frequencies and heterozygosity of the cathepsin, GHRH and LEPR genes
The SNP polymorphisms of the cathepsin CTSB, CTSL and CTSK genes were segregating in the Ukrainian Large White population analysed. Overall, these three SNPs showed extreme allelic frequencies, with major allele frequency (MAF) ranging from 0.02 to 0.09 (Table 2). Major alleles were A for the CTSB g.72A>C SNP, C for the CTSL g.143C>T SNP and G for the CTSK g.15G>A SNP. Consequently, observed heterozygosity (Ho) was low, indicating low genetic diversity at these loci. Expected (He) heterozygosity values were in agreement with observed heterozygosity (Ho), which indicates that these loci are in Hardy–Weinberg equilibrium. The narrow level of genetic diversity of these three loci is also shown by the low polymorphic information content (PIC) of these markers (Table 2). The allele frequencies determined in this study for the Ukrainian Large White pigs were similar to those observed by [7] for CTSB g.72A>C in Italian Large White pigs (MAF = 0.06 for allele C). According to [7], g.72A was also predominant in Landrace, Belgium Landrace, Duroc, Piétrain and Hampshire breeds. Also for CTSL, the results were consistent with previous data indicating the predominance of g.143C allele in Italian Large White, Italian Landrace, Piétrain, Belgian Landrace, Hampshire and Meishan [9]. Similarly, for CTSK, allele g.15G has also been detected as the predominant allele in Italian Large White pig [30].
In contrast to the above, the CTSS G>A polymorphism at the g.171 site was not segregating in Ukrainian Large White pigs as all the animals had genotype GG, although it has been reported to segregate in Italian Duroc, Italian Landrace Hampshire, Belgium Landrace and Piétrain in the range of 0.63–0.95 for the allele g.171G [9, 11]. The absence of segregation might be due to a different origin of Ukrainian Large White which derived from English Large White pigs.
On the other hand, GHRH and LEPR polymorphisms segregated in Ukrainian Large White pigs at close to intermediate frequencies (Table 2). Allele B was the most prevalent for GHRH AluI polymorphism (allelic frequency of 0.73), and allele C was the most prevalent for the LEPR c.1987C>TSNP (allelic frequency of 0.67). A previous study showed that the B allele in GHRH gene was the most prevalent in Large White and Meishan pigs used as first generation of the PiGMap reference families [28], which is in agreement with our results. As seen for the other SNPs, the observed and expected heterozygosity values were very similar. In our case, the PIC of both markers was very high (0.317 and 0.344, respectively), considering that the maximum PIC for a two-allele polymorphism is 0.375. This level of in formativeness is most favourable for undertaking association studies [31].
Analysis of associations between CTSB, CTSL, GHRH and LEPR genes and meat quality characteristics
We have investigated the contribution of mutations in six candidate genes to several muscle attributes related to the quality of meat in Ukrainian Large White pigs. Large White breed is widely used worldwide for efficient meat production, and it is particularly appreciated for the high lean content in prime cuts. Four cathepsin genes (CTSB, CTSL, CTSS, and CTSK) have been investigated in this study. Cathepsins are proteases, which are involved in structural and biochemical changes that take place during post-mortem storage of meat [32]. Large differences in the activities of these enzymes have been detected among pig genotypes [33] with Large White pigs showing a particularly high activity of cathepsin B in biceps femoris. Association analysis between the genotype of the cathepsin genes and meat quality-related traits was performed for CTSB and CTSL (but not for CTSK) loci, as only two pigs were heterozygous for the CTSK polymorphism.
We did not established any significant relationship between the g.72A>C CTSB polymorphism and total protein content, water, ashes, intramuscular fat content and muscle shear force (Table 3). The protein content in the muscles of Ukrainian Large White pigs was in a range of 20.5–21.8 g/100 g of tissue which is slightly higher when compared to the muscle protein level in Large White pigs (19.5–20.0 mg/100 g of tissue) reported by [34]. At the same time, the Ukrainian Large White population had a lower average intramuscular fat content of 1.61 g/100 g of tissue when compared to a commercial Large White cross breed (1.91 g/100 g of tissue [35]). There was a tendency towards a lower muscle pH value at 48 h post-mortem in g.72AA pigs when compared to g.72AC, but these differences were not statistically significant (P = 0.099). The average pH value in our study was 5.53 which is comparable to pH values in purebred Large White and Duroc × Landrace × Large White cross breed (5.88 and 5.57 respectively) [36, 37]. According to the literature, pH values at 48 h are similar or slightly lower when compared to the pH at 24 h. The reason for this is that up to 24 h after slaughter, the glycogen in the muscle is rapidly converted into lactic acid resulting in pH decrease from 6.8–7.3 to 5.4–5.8. During the following 48 h, the meat goes through the process of maturation which is not accompanied by active glycogen conversion and therefore does not result in significant changes in pH values [38].
CTSB polymorphism had been previously studied with regards to growth and fattening traits in Italian Large White pigs [7, 26] showing a significant effect on backfat thickness [27]. These studies did not find effect of CTSB polymorphism on early muscle pH [7, 27]. More encouraging results were obtained in our study on CTSL (g.143C>T genotype, Table 3). On this locus, the total protein content was higher in pigs with g.143CC genotype when compared to CT animals (P = 0.023). In contrast, pigs with g.143CT genotype had a higher meat pH value then that for g.143CC pigs (P = 0.028). This tendency was also reported by [9] in Italian Large White pigs. However, it should be noted that in the present study, pH was measured at 48 h post-mortem whilst in the study by [9], pH was analysed at 2 and 24 h post-mortem which does not allow to make direct comparison.
The GHRH gene was selected for this study because of its relationship with growth and fat deposition, the two traits which have important implications for meat quality. Previous studies proved that injection of GHRH to pregnant sows increases piglets’ weight, both at birth and at weaning, and reduced the time required to reach the market [39, 40].
The present study established associations between GHRH polymorphisms, intramuscular fat content and meat tenderness, assessed as muscle shear force (Table 4). Pigs with GHRH BB genotype had a higher intramuscular fat content when compared to the animals with AB and AA genotypes. The relationship between this GHRH polymorphism and the carcass fat is well established. This can be illustrated by studies on Landrace pigs where the animals with GHRH BB genotype had thicker back fat when compared to AA and AB pigs [14]. Similarly, in an experimental group consisting of Yorkshire, Duroc and Landrace pigs, the animals with AA genotype had a 1.23 % higher meat percentage when compared to the animals with BB genotype [41].
Information available on pure-bred Large White pigs is fragmental and often inconsistent. Thus, although backfat thickness in pure-bred Large White has been reported to be affected by the GHRH genotype [42], the other authors [43] did not find effect of the GHRH/AluI polymorphism on intramuscular fat and lean meat percentage, but consistently showed effect on average back fat thickness in this breed. This discrepancy might be related to the fact that [42] and [43] conducted their studies on different types of fat (subcutaneous and intramuscular fat respectively). Although positive correlation between intramuscular and subcutaneous fat content in pigs has been reported in a number of publications [44], there is an increasing number of studies suggesting that these two fat depots are regulated by different mechanisms and/or different genes [45, 46] and that the mechanisms of fat deposition might be breed-specific.
In the present study, the GHRH AluI polymorphism has also significantly affected meat tenderness. Unexpectedly, our Large White population of AluI-BB pigs, which had a higher intramuscular fat content, exhibited tougher meat (Table 4). The relationship between intramuscular fat content and meat tenderness is very much in dispute [47–49]. The latest reports indicate that in pigs tenderness positively correlates with intramuscular fat content values above 2 % [2], which is above the range of intramuscular fat in most of the animals used in present study. Moreover, intramuscular fat correlates with collagen content [49], which influences the mechanical strength of meat [50] and might explain the contradictory results between tenderness and intramuscular fat content across studies. The GHRH mutation had a significant dominant component for shear force and a high additive component for intramuscular fat content (Table 5). All these results are consistent with the fact that, in Ukrainian Large White pigs, allele A has a significant effect on favourable traits such as lower fat and more tender meat.
The g.1987C>T polymorphism in the LEPR gene also had significant effects on intramuscular fat content and tenderness values in Ukrainian Large White pigs in this study. Animals with TT genotype, had a higher intramuscular fat content (P = 0.02) and, consequently, a lower amount of ashes (P = 0.02) and a tendency to a lower water content (P = 0.22) (Table 6). This is consistent with the data of literature that intramuscular fat content has significant negative relation with moisture and water content [51].
Allele C behaves for this two traits in a complete dominant manner (Table 6), showing no difference between CC and CT genotypes. Accordingly, significant dominant effects were found for ash and fat content along with an additive component that highlights the differences reported in TT pigs (Table 5). The LEPR c.1987C>T mutation involves a L663F amino acid change in the coded protein that could be responsible for a reduction of the receptor function or signalling ability [52]. These data are consistent with results of other authors [17, 53] on Duroc x Iberian crosses and [18] on Iberian x Landraces cross-breed which demonstrated a higher association between the fat content and LEPR c.1987T allele when compared to c.1987C allele. On the other hand, there was a strong underdominant effect of this mutation on mechanical shear force, with CT pigs displaying the lower values (P = 0.02) and, therefore, a more tender meat (Table 6). This mutation has a significant dominant component for muscle shear force but the additive effect was not significant, emphasising the benefit of heterozygous CT animals, which would have, on the whole, more tender meat and lower fat content (Table 5).
To summarise, this study reports new data on allele variance in CTSL, CTSB, CTSS and CTSK genes as well as in GHRH and LEPR in pure-bred Ukrainian Large White pigs. In particular, a medium level of heterozygosity values was established for GHRH and LEPR genes which corresponded to very high levels of informativeness indexes. In contrast, cathepsins CTSL, CTSB and CTSK had a low level of heterozygosity, and CTSS did not segregate in this breed. Association studies demonstrated that intramuscular fat content and tenderness were affected by the allele variance in GHRH and LEPR but not by CTSB and CTSL genes. The GHRH results could be particularly relevant for the production of lean prime cuts as the A allele is associated with both, a lower meat fat content and, most importantly, better tenderness values, which are the two attributes highly regarded by consumers. The effect of this allele is in contrast with the overdominant manner by which the LEPR enhances tenderness and lowers fat content. Results of this study suggest that selective breeding towards GHRH/AA genotype would be particularly useful for improving meat quality characteristics in the production systems involving lean Large White lines with typical intramuscular fat content below 2 %. However, it should be noted that a relatively small number of animals is a limitation of this study and therefore further research and validation of these results on a large population is needed.
References
Wood JD, Richardson RI, Nute GR, Fisher AV, Campo MM, Kasapidou E, Sheard PR, Enser M (2003) Effects of fatty acids on meat quality: a review. Meat Sci 66:21–32
Gao SZ, Zhao SM (2009) Physiology, affecting factors and strategies for control of pig meat intramuscular fat. Recent Pat Food Nutr Agric 1:59–74
Olsson V, Pickova J (2005) The influence of production systems on meat quality with emphasis on pork. Ambio 34:4–5
Switonski M, Stachowiak M, Cieslak J, Bartz M, Gres M (2010) Genetics of fat tissue accumulation in pigs: a comparative approach. J Appl Genet 51(2):153–168
Etherington DJ (1984) The contribution of the proteolytic enzymes to pos-mortem changes in muscle. J Anim Sci 59:1644–1649
Virgili R, Schivazappa C, Parolari G, Soresi-Bordini C, Degni M (1998) Proteases in fresh pork muscle and their influence on bitter taste formation in dry-cured ham. J Food Biochem 22:53–63
Russo V, Fontanesi L, Scotti E, Beretti F, Davoli R, Nanni Costa L, Virgili R, Buttazzoni L (2008) Single nucleotide polymorphisms in several porcine cathepsin genes are associated with growth, carcass, and production traits in Italian Large White pigs. J Anim Sci 86:3300–3314
Dvorakova V, Stupka R, Sprysl M, Citek J, Okrouhla M, Kluzakova E, Brzobohaty L (2011) The effect of missense mutation G.143C>T in the CTSL gene on production traits without the effect on quality of pork meat. Res Pig Breed 5:18–21
Fontanesi L, Speroni C, Buttazzoni L, Scotti E, Nanni Costa L, Davoli R, Russo V (2010) Association between cathepsin L (CTSL) and cathepsin S (CTSS) polymorphisms and meat production and carcass traits in Italian Large White pigs. Meat Sci 85:331–338
Fontanesi L, Speroni C, Buttazzoni L, Scotti E, Dall’Olio S, Nanni Costa L, Davoli R, Russo V (2010b) The insulin-like growth factor 2(IGF2) gene intron3-g.3072G>A polymorphism is not the only Sus scrofa chromosome 2p mutation affecting meat production and carcass traits in pigs: evidence from the effects of a cathepsin D (CTSD) gene polymorphism. J Anim Sci 88(7):2235–2245
Fontanesi L, Speroni C, Buttazzoni L, Scotti E, Dall’Olio S, Davoli R, Russo V (2012) Association between polymorphisms in Cathepsin and cystatin genes with meat production and carcass traits in Italian Duroc pigs: confirmation of the effects of a Cathepsin L (CTSL) gene marker. Mol Biol Rep 39(1):109–115
Ramos AM, Glenn KL, Serenius TV, Stadler KJ, Rothschild MF (2008) Genetic markers for the production of US country hams. J Anim Breed Genet 128:248–257
Piorkowska K, Ropka-Molik K, Eckert R, Tyra M, Zukowski K (2012) The association between polymorphisms of three cathepsins and and economically important traits in pigs raised in Poland. Livest Sci 150:316–323
Pierzchala M, Blicharski T, Kuryl J (2003) Growth rate and carcass quality in pigs as related to genotype at loci POU1F1/RsaI (Pit1/RsaI) and GHRH/AluI. Anim Sci Pap Rep 21(3):159–166
Franco MM, Antunes RC, Silva HD, Goulart LR (2005) Association of PIT1, GH and GHRH polymorphisms with performance and carcass traits in Landrace pigs. J Appl Genet 46(2):195–200
Kovacik A, Bulla J, Trakovicka A, Lieskovska Z, Zitny J (2011) Effect of porcine LEPR polymorphism (Hpaii) on Carcass Traits in Large White A: Landrace Crossbred Pigs. Anim Sci Biothechnol 44(1):260
Munoz G, Alcazar E, Fernandez A, Barragan C, Carrasco A, de Pedro E, Silio L, Sanchez JL, Rodriguez MC (2011) Effects of porcine MC4R and LEPR polymorphisms, gender and Duroc sire line on economic traits in Duroc x Iberian crossbred pigs. Meat Sci 88:169–173
Perez-Montarelo D, Fernandez A, Folch JM, Pena RN, Ovilo C, Rodriguez C (2012) Joint effect of porcine leptin and leptin receptor polymorphisms on productivity and quality traits. Anim Genet 43:805–809
Jennen DGJ, Brings AD, Liu G, Jüngst H, Tholen E, Jonas E, Tesfaye D, Schellander K, Phatsara C (2007) Genetic aspects concerning drip loss and water-holding capacity of porcine meat. J Anim Breed Genet 124(Suppl. 1):2–11
Briggs HM (1983) International Pig Breed Encyclopedia. Elanco Products Company
FAO, Food and Agriculture Organization of the United Nation (2014) Breeding strategies for sustainable management of animal. http://www.fao.org/docrep/012/i1103e.pdf. Accessed 28 Oct 2014
European Convention for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes, Strasbourg, 18.III.1986. http://conventions.coe.int/treaty/en/treaties/html/123.htm
Fontanesi L, Scotti E, Speroni C, Buttazzoni L, Russo V (2011) A selective genotyping approach identifies single nucleotide polymorphisms in porcine chromosome 2 genes associated with production and carcass traits in Italian heavy pigs. Ital J Anim Sci 10(2):e15
Fujii J, Otsu K, Zorzato F, DeLeaon S, Khanna VK, Weiler JE, O’Brien PJ, MacLennan DH (1991) Identification of a mutation in the porcine ryanodine receptor associated with malignant hypothermia. Science 253:448
AOAC (1990) Official methods of analysis, 15th edn. Association of Official Analytical Chemists, Arlington, pp 389–391
Boccard R, Buchter L, Casteels M, Cosentino E, Drangseld E, Hood DE, Joseph RL, MacDougall DB, Rhodes DN, Schoen I, Tinbergen BJ, Touraille C (1981) Procedures for measuring meat quality characteristics in beef production experiments. Report of a Working Group in the Commission of the European Communities (CEC) Beef Production Research Programme. Livest Prod Sci 8:385–397
Russo V, Fontanesi L, Davoli R, Nanni Costa L, Cagnazzo M, Buttazzoni L, Virgili R, Yerle M (2002) Investigation of candidate genes for meat quality in dry-cured ham production: the porcine cathepsin B (CTSB) and cystatin B (CSTB) genes. Anim Genet 33:123–131
Baskin LC, Pomp D (1997) Restriction fragment length polymorphism in amplification products of the porcine growth hormone-releasing hormone gene. J Anim Sci 75:2285
Peakall R (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Fontanesi L, Scotty E, Buttazzoni L, Dall’Olio S, Davoli R, Russo V (2010) A single nucleotide polymorphism in the porcine Cathepsin K (CTSK) gene is associated with back fat thickness and production traits in Italian Duroc pigs. Mol Biol Rep 37:491–495
Hao LL, Yu H, Zhang Y, Sun SC, Liu SC, Zeng YZ, Ali YX, Jiang HZ (2011) Single nucleotide polymorphism analysis of exons 3 and 4 of IGF-1 gene in pigs. Genet Mol Res 10:1689–1695
Flores M, Aristoy MC, Antequera T, Barat JM, Toldrá F (2012) Effect of brine thawing/salting on endogenous enzyme activity and sensory quality of Iberian dry-cured ham. Food Microbiol 29(2):247–254
Hernandez P, Zomeno L, Arino B, Blasco A (2004) Antioxidant, lipolytic and proteolytic enzyme activities in pork meat from different genotypes. Meat Sci 66(3):525–529
Choi YM, Nam KW, Choe JH, Ryu YC, Wick MP, Lee K, Kim BC (2013) Growth, carcass, fiber type, and meat quality characteristic in Large White pigs with different liveweights. Livest Sci 155:123–129
Bessa RJB, Hughes RA, Jeronimo E, Moreira OC, Prates JAM, Doran O (2014) Effect of pig breed and dietary protein level on selected fatty acids and stearoyl-coenzyme A desaturase protein expression in longissimus muscle and subcutaneous fat. J Anim Sci 91:4540–4546
Renaudeau D, Mourot J (2007) A comparison of carcass and meat quality characteristics of Creole and Large White pigs slaughtered at 90 kg BW. Meat Sci 76:165–171
Ma X, Zheng C, Hu Y, Wang L, Yang X, Jiang Z (2015) Dietary L-arginine supplementation affects the skeletal longissimus muscle proteome in finishing pigs. PLoS One 10(1):e0117294
Desai BB (2000) Handbook of nutrition and diet., Series: food science and technology. Book 104CRC Press, Boca Raton
Khan AS, Bodles-Brakhop AM, Fiorotto ML, Draghia-Akli R (2009) Effect of maternal plasmid GHRH treatment on offspring growth. Vaccine 28(8):1905–1910
Draghia-Akli R, Fiorotto ML (2004) A new plasmid-mediated approach to supplement somatotropin production in pigs. J Anim Sci 82(E-Suppl):E264–E269
Cho ES, Park DH, Kim BW, Jung WY, Kwon EJ, Kim CW (2009) Association of GHRH, H-FABP and MYOG polymorphisms with economic traits in pigs. Asian Australas J Anim Sci 3:307–312
Blicharski T, Zak G, Pierzchala M, Eckert R (2003) Preliminary studies on possibility of estimating pig belly lean percentage based on slaughter traits measured at a meat plant. Ann Anim Sci 2:333–344
Piorkowska K, Ropka-Molik K, Oczkowicz M, Rozycki M, Zukowski K (2013) Association study of PIT and GHRH SNPs with economically important traits in pigs of three breeds reared in Poland. Anim Sci Pap Rep 31(4):303–314
Ros-Freixedes R, Reixach J, Bosch L, Tor M, Estany J (2013) Response to selection for decreased backfat thickness at restrained intramuscular fat content in Duroc pigs. J Anim Sci 91(8):3514–3521
Hocquette JF, Gondret F, Baeza E, Medale F, Jurie C, Pethick DW (2010) Intramuscular fat content in meat-producing animals: development, genetic and nutritional control, and identification of putative markers. Animal 4:303–319
Hernandez-Sanchez J, Amills M, Pena RN, Mercade A, Manunza A, Quintanilla R (2013) Genomic architecture of heritability and genetic correlations for intramuscular and back fat contents in Duroc pigs. J Anim Sci 91:623–632
Hovenier R, Kanis E, Verhoeven JA (1993) Repeatability of taste panel tenderness scores and their relationships to objective pig meat quality traits. J Anim Sci 71(8):2018–2025
Fortin A, Robertson WM, Tong AK (2005) The eating quality of Canadian pork and its relationship with intramuscular fat. Meat Sci 69:297–305
Suzuki K, Irie M, Kadowaki H, Shibata T, Kumagai M, Nishida A (2005) Genetic parameter estimates of meat quality traits in Duroc pigs selected for average daily gain, longissimus muscle area, backfat thickness, and intramuscular fat content. J Anim Sci 83(9):2058–2065
Nishimura T (2015) Role of extracellular matrix in development of skeletal muscle and postmortem aging of meat. Meat Sci 109:48–55
Kim JH, Seong PN, Cho SH, Park BY, Hah KH, Yu LH, Lim DG, Hwang IH, Kim DH, Lee JM, Ahn CV (2008) Characterization of nutritional value for twenty-one pork muscles. Asian-Aust J Anim Sci 21:138–143
Ovilo C, Fernandez A, Noguera JL, Barragan C, Leton R, Rodriguez C, Mercade A, Alves E, Folch JM, Varona L, Toro M (2005) Fine mapping of porcine chromosome 6 QTL and LEPR effects on body composition in multiple generations of an Iberian by Landrace intercross. Genet Res 85:57–67
Munoz G, Ovilo C, Silio L, Tomas A, Noguera JL, Rodriguez MC (2009) Single- and joint-population analyses of two experimental pig crosses to confirm quantitative trait loci on Sus scrofa chromosome 6 and leptin receptor effects on fatness and growth traits. J Anim Sci 87:459–468
Acknowledgments
This research was supported by the National Academy of Agricultural Sciences, Ukraine (No DP 0111U004024), EU Marie Curie Grant (IRSES No 246760) and the UK Biotechnology and Biological Sciences Research Council (BBSRC) grant (BB/L024268/1).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Balatsky, V., Bankovska, I., Pena, R.N. et al. Polymorphisms of the porcine cathepsins, growth hormone-releasing hormone and leptin receptor genes and their association with meat quality traits in Ukrainian Large White breed. Mol Biol Rep 43, 517–526 (2016). https://doi.org/10.1007/s11033-016-3977-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-016-3977-z