Abstract
A new cysteine peptidase (Granulosain I) was isolated from ripe fruits of Solanum granuloso-leprosum Dunal (Solanaceae) by means of precipitation with organic solvent and cation exchange chromatography. The enzyme showed a single band by SDS-PAGE, its molecular mass was 24,746 Da (MALDI-TOF/MS) and its isoelectric point was higher than 9.3. It showed maximum activity (more than 90%) in the pH range 7–8.6. Granulosain I was completely inhibited by E-64 and activated by the addition of cysteine or 2-mercaptoethanol, confirming its cysteinic nature. The kinetic studies carried out with PFLNA as substrate, showed an affinity (Km 0.6 mM) slightly lower than those of other known plant cysteine proteases (papain and bromelain). The N-terminal sequence of granulosain I (DRLPASVDWRGKGVLVLVKNQGQC) exhibited a close homology with other cysteine proteases belonging to the C1A family.
Similar content being viewed by others
1 Introduction
The cysteine proteases of clan CA, family C1 (the papain family), are a group of enzymes identified in bacteria, yeast, animals, and plants, which play an important role in intracellular protein degradation [1]. These enzymes share not only their general architecture but also the micro-arrangement of the catalytic residues Cys 25, His 159 and Asn 175 (according to the papain numbering). The ionized state of the nucleophilic cysteine residue in the active site is independent of substrate binding, making these and other cysteine proteases active a priori [2].
The family of papain-type proteinases is the most thoroughly investigated among all the cysteine proteinases. Papain is characterized by a two-domain structure with the active site between the domains. These enzymes are usually synthesized as inactive or less active precursors. Activation takes place by limited intra- or intermolecular proteolysis [3]. To date, the amino acid sequences of more than 50 papain-like and 15 legumain-like plant proteinases have been established [4]. They are stored in the vacuole or the lysosome, or are externally secreted. Among the cysteine proteases, the H and L cathepsins have been widely studied in mammals and, more recently, in plants [5].
Plant proteinases of this class are mainly used to mobilize storage proteins in seeds. Protein bodies of seeds contain both storage proteins and protease precursors. The latter become activated after germination and start degradation of the stored proteins [6]. The activities of cysteine proteinases respond dramatically to different internal and external stimuli and in some cases they rise to 90% of total proteolytic activity. They are involved in protein maturation, degradation and protein rebuilt in response to different external stimuli and they also play a house-keeping function to remove abnormal misfolded proteins [7].
Some of these enzymes have medical significance because they are resorbed in the gut as active enzymes and may elicit an immune response [8, 9].
A number of these proteinases have an essential role in processes used by the modern food and feed industry, to produce a wide range of products for human and animal consumption. They have been exploited commercially in the food industry for meat tenderizing (partially separating connective tissues), brewing (to solubilize grain proteins and stabilize beer), and cookie baking (to improve crispness), as well as the production of protein hydrolysates [10]. Protein hydrolysates are currently employed to produce many foods in which enzymes can replace potentially carcinogenic or otherwise harmful chemicals. They also have applications in tanning, leather and textile industries, to remove hair, wool, and to soften skins [11].
Our groups are dedicated to the search for new sources of plant proteolytic enzymes, their purification and biochemical characterization. Some of us [12, 13] have studied stabilization by adsorption and are exploring potential biotechnological applications. Here we report the characterization of Granulosain I, the major protease purified from crude extracts of ripe fruits of Solanum granuloso-leprosum (Solanaceae).
In contrast to the rest of this family, S. granuloso-leprosum is a tree growing 4–6 m tall, whereas most other members are plants or shrubs, 1 or 2 m tall. Another distinguishing characteristic with the rest of this family, is that the S. granuloso-leprosum is an arboreal constitution (being able to reach 4–6 m high) whereas the majority of the members have constitution type kills or shrub (1 or 2 m high).
The Solanaceae are also the third most important plant taxon economically and the most valuable in terms of vegetable crops, and are the most variable of crops species in terms of agricultural utility, as it includes the tuber-bearing potato, a number of fruit-bearing vegetables (tomato, eggplant, peppers), ornamental plants (petunias, Nicotiana), plants with edible leaves (Solanum aethiopicum, S. macrocarpon) and medicinal plants (e.g. Datura, Capsicum) [14].
2 Material and Methods
2.1 Chemicals
AMPSO4, CAPS, casein, Coomasie Brillant Blue R-250 and G-250, cysteine, E-64, EDTA, MOPS, PFLNA, PMSF, PVDF membrane, TAPS, Tris base and glycine were purchased from Sigma Chemical Company (St. Louis, MO, USA). Acrylamide, bisacrylamide, and low-range molecular weight standards were obtained from Bio-Rad (Hercules, CA, USA) CM-Sepharose Fast Flow and Pharmalyte 3-10 were purchased from Pharmacia (Biotech. Uppsala, Sweden). All other chemical were obtained from commercial sources and were of the highest available purity.
2.2 Plant Material
Ripe fresh fruits from S. granuloso-leprosum Dunal [syn = S. verbascifolium var. auriculatum (Ait.) O.K.], Solanaceae, were collected in January from wild trees that grow in Montevideo city. The plant (a shrub or small tree) grows in northeastern Argentina, Uruguay and Brazil and the fruits are orange berries, about 1–2 cm long.
2.3 Crude Extract Preparation
A crude extract was prepared by grinding fresh mature fruits, previously washed and dried, in a mortar. Homogenates were filtered through a piece of gauze folded in two to remove plant debris, and then centrifuged for 15 min at 6,654g. The supernatant (crude extract) was stored at −20 °C for further analysis.
2.4 Preliminary Purification of Crude Extract
The crude extract was treated with four volumes of cold (−20 °C) acetone with gentle agitation and left to settle for 20 min before centrifugation at 6,654g for 30 min. The final acetone precipitate (RAP) was redissolved with one volume of 50 mM buffer Tris–HCl pH 7.5 [15].
2.5 Protein Content Determination
Proteins present in the crude extract and in the partial purified fractions were measured according to Bradford’s method [16] using bovine seroalbumin as the standard. The protein content of chromatographic fractions was estimated by measurement of absorbance at 280 nm.
2.6 Proteolytic Activity Determination
Casein was the substrate used in most cases. The reaction mixture contained 0.1 mL of crude extract and 1.1 mL of 1% casein in a 0.1 M Tris–HCl buffer (pH 7.5) with 15 mM cysteine (final concentration). The reaction was carried out at 37 °C and stopped 20 min later by adding 1.8 mL of 5% (w/v) trichloroacetic acid (TCA). Each test tube was centrifuged at 6,000g for 30 min and the absorbance of the supernatant was measured at 280 nm. One caseinolytic unit (Ucas) was defined as the amount of protease that produces an increment of one absorbance unit per minute in the assay conditions [17].
Azocasein was the substrate used in titration assays with E-64. The reaction mixture containing 340 μL of an appropriate dilution of the enzyme preparation, 340 μL of azocasein solution (1% w/v in distiller water) and 340 μL of buffer (0.1 M Tris–HCl buffer, pH 7.5) was incubated for 20 min at 37 °C. The reaction was stopped by adding 340 μL of TCA (10% w/v in distilled water). After centrifuging for 20 min at 20,600g, absorbance was measured at 337 nm. In this case, one enzymatic unit (Uazo) was defined as the quantity of enzyme required to increase by one absorbance unit the absorbance value at 337 nm per minute in the assay conditions [18].
2.7 Cation-Exchange Chromatography
The RAP (redissolved acetone precipitate) was purified by cation exchange chromatography on a column (Pharmacia XK 16/40, with AK16 adaptors) packed with CM-Sepharose Fast Flow, equilibrated and washed with 50 mM Tris–HCl buffer (pH 7.5). Chromatography was carried out in FPLC equipment (Pharmacia) by washing with the equilibrating buffer and further elution of the bound material with a linear sodium chloride gradient (0–0.3 M) in the same starting buffer at a flow rate of 2 mL/min. The protein content of the chromatographic fractions was monitored spectrophotometrically by measuring absorption at 280 nm. The eluted fractions were assayed for caseinolytic activity, and those showing proteolytic activity were pooled and stored at −20 °C for further studies.
2.8 Optimum pH
Caseinolytic activity was measured at 37 °C at different pH values (6–10) using 10 mM sodium salts of the following “Good” buffers: MES, MOPS, TAPS, AMPSO and CAPS [19].
2.9 Effect of Activators
The effect of activity enhancers was determined by measuring the caseinolytic activity in 0.1 M Tris buffer pH 7.5 after preincubating the sample with the activator at 37 °C for 30 min. The activators assayed were β-mercaptoethanol and cysteine (5, 10, 15, 20, 30 and 50 mM final concentration). The activity was compared with the enzyme without the addition of inhibitors or activators.
2.10 Titration of the Active Site with E-64
Titration of the active site was performed as described by Salvesen and Nagase [20], with some modifications. The enzyme (0.6 μM) was preincubated with the activation buffer (50 mM Tris–HCl pH 7.5, containing 15 mM cysteine). Fractions (0.75 μL) were incubated with 25 μL of different concentrations (0–2 μM) of E-64 for 30 min at 37 °C and the residual azocaseinolytic activity was measured. Enzyme concentration was established by determining both protein content (Bradford method) and molecular mass value (mass spectrometry).
2.11 Isoelectric Focusing (IEF) and Zymogram
Isoelectric focusing (IEF) was carried out in 5% polyacrylamide gels containing a broad pH range of ampholytes (Biolyte 3-10, Bio-Rad) in a Mini IEF Cell (Model 111, Bio-Rad). Samples were precipitated with four volumes of cold (−20 °C) acetone and redissolved twice in deionized water. About 1–10 μg of protein was loaded in each case. Focusing was carried out under constant voltage conditions in a stepped procedure: 100 V for 15 min, 200 V for the following 15 min, and 450 V for the last 60 min.
One of the gels was fixed and stained with Coomassie Brilliant Blue R-250, while the other, unstained, was put in contact with an agarose gel of the same dimensions preequilibrated with 1% casein solution for 20 min at 56 °C, in order to detect bands with proteolytic activity [21]. After incubation, the agarose gel was dehydrated and stained with Coomassie Brilliant Blue R-250.
2.12 Electrophoresis
After each purification step, the fraction was tested by SDS-PAGE in 12.5% polyacrylamide gels [22]. Current was kept constant at 30 V during stacking and then increased to 90 V and kept constant for the resolving gel. The protein bands were visualized by staining with Coomassie Brilliant Blue R-250.
2.13 Mass Spectrometry
Purity, and the molecular weight of the contents of chromatographic fractions, were determined by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF/MS).
MALDI-TOF mass spectra were obtained using a Bruker Daltonics® model Ultraflex spectrometer equipped with a pulsed nitrogen laser (337 nm), in linear positive ion mode, using a 20-kV acceleration voltage. Samples were prepared by mixing equal volumes of a saturated solution of the matrix (sinapinic acid) in 0.1% TFA (aq.) acetonitrile (2:1), and the protein solution. From this mixture, 0.5 μL was spotted on to a simple slide and allowed to evaporate to dryness. Bovine trypsinogen was used for internal calibration.
2.14 Amidolytic Activity and Kinetic Parameters
Amidolytic activity was determined using PFLNA as substrate at 45 °C by spectrophotometric measurement of absorbance at 410 nm, according to Filippova et al. [23]. The reaction mixture contained 1.5 mL of 0.1 M phosphate buffer pH 7.5, containing 0.3 M KCl, 0.1 mM EDTA and 3 mM DTT, 180 μL of substrate (0.025–0.5 mM final concentration) and 120 μL of enzyme. An arbitrary enzyme activity unit (UPFLNA) was defined as the amount of protease that released 1 μmol of p-nitroaniline per min in the assay conditions. The kinetic parameters (Vm, Km y Kcat) of granulosain I were calculated using linear regression analysis by means of Lineweaver-Burke, Wolf-Hanes y Eadie-Hofstee plots.
2.15 N-terminal Sequence
The protein band in the SDS-PAGE of the purified protease, granulosain I, was transferred onto a PVDF membrane (Millipore) and washed several times with deionized water. The N-terminal sequence was determined by Edman’s automated degradation using a Beckman LF3000 protein sequencer equipped with a System Gold (Beckman) PTH-amino acid analyzer. Protein homology searches were performed using the Blast network service [24].
3 Results
3.1 Purification of Crude Extract
The crude extract used for the isolation of granulosain I was obtained from ripe fruits of S. granuloso-leprosum.
Previous studies carried out on this extract found it had significant proteolytic activity (8 Ucas/mg) and determined the physicochemical parameters in which this enzymatic preparation showed maximum activity and stability [25].
The purification of proteases from the crude extract was achieved by a two-step procedure. The first step was the treatment of the crude extract with four volumes of cold acetone (−20 °C) to eliminate soluble carbohydrates as well as phenolic compounds, which could oxidize and react irreversibly with the proteins present in the homogenate. The redissolved acetone precipitate (RAP) contained 89% of the proteins and 76% of the total caseinolytic activity present in the crude extract.
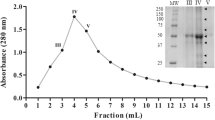
The second purification step was column chromatography of the RAP using a CM-Sepharose FF column in a FPLC system. The elution profile is shown in Fig. 1. The unbound eluate exhibited low activity toward casein, but the two fractions eluted after the application of the linear NaCl gradient it were both proteolytically active. The second peak (P II), which eluted at 0.25 M NaCl, showed higher specific activity (10 Ucas/mg) and was therefore selected for further study.
3.2 Biochemical Characterization of Granulosain I
SDS-PAGE analysis of peak II fractions showed a single homogeneous protein band. Mass spectrometry confirmed the homogeneity of this protein, its molecular mass (Mr) was 24,746 Da (Figs. 2 and 3).
According to nomenclature recommendations for proteases [26], a protease of peak II was named Granulosain I.
Isoelectric focusing and specific revealed for proteases (zymogram) showed that granulosain I had a pI higher than 9.3 (Fig. 2).
The pH range in which granulosain I showed high caseinolytic activity (greater than 90%) was 7.0–8.6 (Fig. 3).
The enzyme was completely inhibited by E-64 and iodoacetic acid. This behaviour is similar to that of the C1A family of cysteine proteases [12].
The effect of increasing concentrations of cysteine and 2-mercaptoethanol on granulosain I activity was studied. It was observed that both activators increased the activity in to a similar extent. In the case of cysteine the concentration that produced the greatest increment in activity (200%) was 15 mM, while for 2-mercaptoethanol the concentration that produced the highest activation level (200%) was 30 mM (Fig. 4).
In active site titration with E-64, when activity was plotted against E-64 concentration, a straight line was obtained, which when extrapolated intersected the abscissa at 1.8 μM E-64, corresponding to 45% of total enzyme (Fig. 5).
3.3 Kinetic Parameters
The amidolytic activity and the main kinetic parameters (Km, Vm, Kcat and Kcat/Km) of granulosain I were determined using PFLNA as substrate which has been extensively used to evaluate the activity of plant cysteine proteinases like papain, bromelain and ficin.
The values obtained for granulosain I was shown in Table 1.
3.4 Sequence Analysis
The N-terminal aminoacid sequence obtained for granulosain I (DRLPASVDWRGKGVLVLVKNQGQC) was compared with sequences of other plant proteases (Table 2).
4 Discussion
Supported in a marked interest by the available regional vegetable biodiversity, it is searched to obtain proteolytic catalysts that allow their application in different biotechnological processes, as well as the development of new undertaking.
Studies previously realized on ripe fruits extract of S. granuloso-leprosum (Solanaceae), demonstrated a significant and unusual proteolytic activity, for this vegetable family.
The Solanaceae family includes about 85 genera and 3000 species [43] but in spite of being an abundant and widely distributed family, to our knowledge there have been no reports of the isolation and biochemical and structural characterization of any proteolytic enzymes belonging to it. The majority of the existing reports come from physiological studies or gene expression from these enzymes under conditions of stress or pathogen attack [28, 44].
In this work we emphasized the functional, structural and enzymologic classification of the isolated protease.
Through simple two step procedure, was obtained a fraction with high specific activity and purity, proved by SDS-PAGE and MALDI-TOF/MS (Figs. 6 and 7) and named Granulosain I.
SDS-PAGE on 12.5% polyacrylamide gel. 10 μg of protein of crude extract and purified enzyme was loaded onto the gel. Lane 1: crude extract; lanes 2 and 5: MW standard markers: Myosin (210 kDa), BSA (78 kDa), glutamate dehydrogenase (55 kDa), alcohol hydrogenase (45 kDa), carbonic anhydrase (34 kDa), myoglobin (23 kDa), lysozyme (16 kDa), and aprotinin (7 kDa) ; Lane 3: redissolved acetone precipitate; Lane 4: granulosain I
The molecular mass of Granulosain I obtained by mass spectrometry was 24,746 Da. This value is close to those reported for most of the plant cysteine proteinases of the C1A family, like papain (23,800 Da), fruit bromelain (25,000 Da), Ficinain (25,000 Da) and chymopapain (24,400 Da) [45].
Isoelectric focusing and zymogram demonstrated the basic characteristic of granulosain I (pI higher than 9.3, Fig. 2), in common with other plant peptidases. This is evidence that in the enzyme’s native structure, the most external amino acids are basic.
The optimal pH range (7.0–8.6) is consistent with that of most cysteine proteases belonged to the papain family. In this pH range, these enzymes have a monoprotonated thiol-imidazole bond (Cys-His) which is essential for catalytic activity [46]. It might therefore be inferred that granulosain I has a similar mechanism to the cysteine proteases of the C1A family.
The complete inhibition of proteolytic activity when granulosain I was preincubated with E-64 or Iodoacetamide proved that the enzyme’s active site contains sulfhydryl residues that are essential for its catalytic activity.
E-64 has broad specificity and a fast reaction mechanism for papain family members. It is virtually ideal as an active-site titrant, because it has been found to react with the sulfhydryl groups of active site on an equimolecular basis [20].
In the titration assay, only the 45% of total enzyme was active.
Reduction of proteolytic activity during the purification steps could be related to denaturalization of protease structure or autolysis events.
A prior reversible inhibition with HgCl2 of enzyme prevents these events (Data not shown).
The activator effect of increasing concentrations of cysteine and 2-mercaptoethanol confirmed the cysteinic nature of the active site and the dependence of catalytic activity on sulfhydryl groups. It also demonstrated that the enzyme required reduced conditions for activity.
The specificity subsite predominant in most peptidases of subfamily C1A is S2, which commonly displays a preference for occupation by a bulky hydrophobic side chain, and not a charged one [1]. The amidolytic activity as well as the main kinetic parameters (Km, Vm, Kcat and Kcat/km) were determined using PFLNA as substrate (Table 1), which has been extensively used to evaluate the activity of plant cysteine proteinases like papain, bromelain and ficin. (Phenylalanine in P2 position which give thiol proteinases specificities).
The Km value obtained for granulosain I (0.6 mM), showed a lower affinity for PFLNA than papain (0.34 mM), bromelain (0.30 mM) and ficin (0.43 mM) [23].
It is probable that a different spatial disposition of the active site amino acids of granulosain I compared with the active sites of papain, bromelain and ficin, determines its differential affinity for the substrate.
The N-terminal sequence of granulosain I (DRLPASVDWRGKGVLVLVKNQGQC) showed two highly conserved regions (DWR and RNQG) present in most peptidases belonging to the C1A family. Though in granulosain I the RNQG region was changed to KNQG, this is a conservative change (both basic residues, Arg to Lys). Granulosain I exhibited a high degree of identity (greater than 70%) with other plant cysteine proteases, especially with those found in the Solanaceae family (Solanum tuberosum, Lycopersicum esculentum) and with the mammalian cathepsins. These proteases share their general architecture as well as also the micro-arrangement of the catalytic triad (Cys 25, His 159 and Asn175, according to papain numbering). The presence of a Gln residue in the 21 position is essential for catalytic activity as it helps the catalytic triad to form the “oxoanion hole”.
Little is known about the physiological functions of these peptidase. In plants, for example, they have been associated with the mobilization of nutrients and defence against pathogens [3, 28, 47]. Many of these enzymes have important applications in medicine and biotechnology.
In conclusion, the biochemical characterization (MW, IEF, pH and inhibition studies) shows that granulosain I is a cysteine proteinase belonging to the CA1 family. The kinetic studies as well as the N-terminal sequence comparison gives granulosain I a differential identity within this family.
This studies defining optimum working conditions, stability to storage and temperature, size and electrophoretic characteristics will allow us to develop guidelines for the use of these enzymes in biotechnological processes.
Abbreviations
- AMPSO:
-
N-(1,1-Dimethyl-2-hydroxyethyl)-3-amino-2-hydroxypropanesulfonic acid
- CAPS:
-
3-(Cyclohexylamino)-1-propanesulfonic acid
- E-64:
-
trans-Epoxysuccinyl-l-leucyl-amido(4-guanidino)butane
- EDTA:
-
Ethylendiaminetetraacetic acid
- MALDI-TOF/MS:
-
Matrix assisted laser desorption ionization time of flight mass spectrometry
- MES:
-
2-Morpholinoethanesulfonic acid
- MOPS:
-
3-(N-Morpholino) propanesulfonic acid
- PFLNA:
-
pGlu-Phe-Leu p-nitroanilide
- PMSF:
-
Phenylmethanesulfonyl fluoride
- PVDF:
-
Polyvinylidene difluoride
- RAP:
-
Redissolved acetone precipitate
- TAPS:
-
N-Tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid
References
Rawlings NM, Barrett AJ (2004) In: Barrett AJ, Rawlings NM, Woessner JF (eds) Handbook of proteolytic enzymes, 2nd edn., vol. II. Academic Press, London, pp 1051–1071
Wiederanders B (2003) Acta Biochimica Polonica 50:691–713
van der Hoorn RAL, Leeuwenburgh MA, Bogyo M, Joosten MHAJ, Peck SC (2004) Plant Physiol 135:1170–1178
Fischer J, Becker C, Hillmer S, Horstmann C, Neubohn B, Schlereth A, Senyuk V, Shutov A, Muntz K (2000) Plant Mol Biol 43:83–101
Ueda T, Seo S, Ohashi Y, Hashimoto J (2000) Plant Mol Biol 44:649–657
Schlereth A, Standhardt D, Mock HP, Muntz K (2001) Planta 212:718–727
Wisniewski K, Zagdanska B (2001). J Exp Bot 52:1455–1463
Furmonaviciene R, Sewell HF, Shakib F (2000) Clin Exp Allergy 30:1307–1313
Nettis E, Napoli G, Ferrannini A, Tursi A (2001) Allergy 56:257–258
Guadix A, Guadix EM, Páez-Dueñas MP, González-Tello P, Camacho F (2000) Ars Pharmaceutica 41:79–89
Uhlig H (1998) Industrial enzymes and their applications. Wiley, New York, pp 146–147
Vallés D, Furtado S, Villadóniga C, Cantera AMB (2004a) Int J Biotechnol 6:346–360
Vallés D, Furtado S, Cantera AMB (2007) Enzyme Microbial Technol 40:409–407
Bohs L, Olmstead RG (1997) Syst Bot 22:5–17
Scopes, RK (1994) In: Robert CG (ed) Protein purification: principles and practice, 3rd edn, vol. 96. Pringer-Verlag NY Inc, New York, pp 317–321
Bradford MB (1976) Anal Biochem 72:248–254
Bruno MA, Pardo MF, Caffini NO, López LMI (2003) J Protein Chem 22:127–134
Castro S, Vázquez D, Cantera AMB (1996) Int Dairy J 5:1–10
Good NE, Izawa S (1972) Meth Enzymol 24:53–68
Salvesen GS, Nagase, H (2001) Inhibition of proteolytic enzymes. In: Beynon R, Bond JS (eds) Proteolytic enzymes, 2nd edn. Oxford University Press, Oxford, pp 105–128
Westergaar JL, Hackbarth C, Treuhaft MW, Roberts RC (1980) J Immunol Meth 34:167–175
Laemmli UK (1970) Nature 227:680–685
Filippova IYu, Lysogorskaya EN, Oksenoit ES, Rudenskaya GN, Stepanov VM (1984) Anal Biochem 143:293–297
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Nucleic Acids Res 25:3389–3342
Vallés D, Bruno MA, González M, López LMI, Cantera AMB, Caffini NO (2004b) Biocell 28:72
Barret AJ (2001) Proteolytic enzymes: nomenclature and classification. In: Beynon R, Bond JS (eds) Proteolytic enzymes, 2nd edn. Oxford University Press, Oxford (UK), pp 1–20
Muller WE, Krasko A, Le Pennec G, Schroder HC (2003) Microsc Res Tech 62:368–377
Avrova AO, Stewart HE, De Jong WD, Heilbronn J, Lyon GD, Birch PR (1999) Mol Plant Microbe Interact 12:1114–1119
Jones ML, Chaffin GS, Eason JR, Clark DG (2005) J Exp Bot 56:2733–2744
Urbich C, Heeschen C, Aicher A, Sasaki K, Bruhl T, Farhadi MR, Vajkoczy P, Hofmann WK, Peters C, Pennacchio LA, Abolmaali ND, Chavakis E, Reinheckel T, Zeiher AM, Dimmeler S (2005) Nat Med 11:206–213
Guiliano DB, Hong X, McKerrow JH, Blaxter ML, Oksov Y, Liu J, Ghedin E, Lustigman S (2004) Mol Biochem Parasitol 136:227–242
Schaffer MA, Fischer RL (1988) Plant Physiol 87:431–436
Lers A, Burd S, Sonego L, Khalchitski A, Lomaniec E (1998) Plant Physiol 116:1193
Morcelle del Valle SR, Trejo SA, Canals F, Avilés FX, Priolo NS (2004) Protein J 23:205–215
Trejo SA, López LMI, Cimino CV, Caffini NO, Natalucci CL (2001) J Protein Chem 20:445–453
The Rice Chromosomes 11 and 12 Sequencing Consortia (2005) BMC Biol 3:20
Ling JQ, Kojima T, Shiraiwa M, Takahara H (2003) Biochim Biophys Acta 1627:129–139
Asamizu E, Sato S, Kaneko T, Nakamura Y, Kotani H, Miyajima N, Tabata S (1998) DNA Res 5:379–391
Taylor MA, Al-Sheikh M, Revell DF, Sumner IG, Connerton IF (1999) Plant Sci 145:41–47
Sequeiros C, Torres MJ, Trejo SA, Esteves JL, Natalucci CL, Lopez LMI (2005) Protein J 24:445–453
Liggieri C, Arribére M, Trejo S, Canals F, Avilés F, Priolo N. (2004) Protein J 23:403–411
Kruger J, Thomas CM, Golstein C, Dixon MS, Smoker M, Tang S, Mulder L, Jones JD (2002) Science 296:744–747
Burkart A (1979) Dicotiledóneas metaclamídeas. In: Flora ilustrada de Entre Ríos Vol.6 parte V, Cientific colection of I.N.T.A, Buenos Aires, pp 346, 347, 358–360
Chen H-J, Huang D-J, Hou W-C, Liu J-S, Lin Y-H (2006) J Plant Physiol 163:863–876
Beers EP, Jones AM, Dickerman AW (2004) Phytochemistry 65:43–58
Polgár L (1973) Eur J Biochem 33:104–109
Grudkowska M, Zagdanska B (2004) Biochim Polonica 51:609–624
Acknowledgments
This work was supported by grants from CYTED (Project IV.22) and PEDECIBA (Chemistry area). Amino acid sequence and mass spectrometry determinations were done at the Institut de Biomedicina y Biotecnología, Universitat Autónoma de Barcelona, Spain. Dr. Valerie Dee undertook linguistic revision of this paper.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Vallés, D., Bruno, M., López, L.M.I. et al. Granulosain I, a Cysteine Protease Isolated from Ripe Fruits of Solanum granuloso -leprosum (Solanaceae). Protein J 27, 267–275 (2008). https://doi.org/10.1007/s10930-008-9133-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-008-9133-4