Abstract
Rice (Oryza sativa L.) leaf color mutants are excellent models for studying chlorophyll biosynthesis and chloroplast development. In this study, we isolated a stable genetic white and lesion mimic leaf1 (wlml1) mutant from an ethyl methanesulfonate (EMS)-mutagenized population of the indica cultivar TN1. Compared with wild-type TN1, the wlml1 mutant had lower contents of chlorophyll and carotenoids, altered chloroplast ultrastructure, and altered regulation of genes associated with chlorophyll metabolism and chloroplast development. In addition, lesions formed on the leaves of wlml1 plants grown at 20 °C and genes related to disease resistance and antioxidant functions were up-regulated; by contrast, the mutant phenotype was partially suppressed at 28 °C. These findings indicated that WLML1 might play a role in chlorophyll metabolism and chloroplast development, as well as in biotic and abiotic stress responses. Genetic analysis showed that WLML1 was controlled by a recessive nuclear gene, and map-based cloning delimited WLML1 to a 159.7-kb region on chromosome 4 that includes 30 putative open reading frames. Based on these findings, the wlml1 mutant will be a good genetic material for further studies on chlorophyll metabolism and stress responses in rice.
Similar content being viewed by others
Introduction
Leaves are the main photosynthetic organs in plants, and changes in leaf color occur during growth and development, including increases in chlorophyll contents on de-etiolation and decreases in chlorophyll during senescence. The study of mutations affecting leaf color can provided insight into these processes. Most of the existing rice mutants displaying altered leaf color are due to mutations in genes related to chloroplast development or metabolism (Zhang and Gao 2009; Yang et al. 2016, Su et al. 2017). These rice leaf color mutations result in decrease of photosynthetic efficiency and crop production (Ye et al. 2017; Ge et al. 2015), and in severe cases can even lead to plant death (Wang et al. 2018). Recently, leaf color mutants have become useful models for studying physiological and metabolic processes such as chlorophyll metabolism, leaf color breeding, disease resistance, and chloroplast structure, function and development. Leaf color mutants also provide useful visible markers for monitoring self-pollination in male-sterile lines and maintaining the purity of genetic stocks. Therefore, it is important to map and clone rice leaf color genes, and investigate how to regulate leaf color in rice. This information will provide a theoretical basis for crop breeding with high photosynthetic efficiency.
Leaf color mutants have been identified in several species, including rice (Hu et al. 1981), soybean (Palmer and Rodriguez de Cianzio 1985), maize (Lin and Yu 1995), barley (Rudoi and Shcherbakov 1998), and wheat (Marco et al. 1989). Leaf color mutants display a variety of leaf colors and patterns including light green, yellow, albino, purple, spots, zebra, altered shades of green, and reversible whitening (Li et al. 2018). The observed changes in leaf color reflect the changes in the type and contents of pigment within the leaves, such as chlorophyll, carotene, and anthocyanin. Chlorophyll plays important roles in determining leaf color as well as capturing and converting light energy into chemical energy (Tanaka and Tanaka 2006; Fromme et al. 2003). Most of the chlorophyll biosynthetic pathway has been determined. It begins with the formation of glutamyl-tRNA, followed by 15 enzymatic reactions involving at least 27 enzymes in higher plants (Liu et al. 2012). Seven chlorophyll biosynthesis genes have been identified in rice: magnesium-chelatase ChlD subunit (OsCHLD), magnesium-chelatase ChlI subunit (OsCHLI), magnesium-chelatase ChlH subunit (OsCHLH), chlorophyll synthase (OsCHLG), divinyl reductase gene (OsDVR), protochlorophyllide oxidoreductase (OsPOR), and chlorophyll a oxygenase (OsCAO) (Nagata et al. 2004). Mutations in these genes affect chlorophyll synthesis, resulting in changes in leaf color. For example, dvr mutants exhibit yellowish-green leaves and stunted plant growth (Wang et al. 2010). Non-yellow coloring 1 (NYC1), which encodes a chlorophyll b reductase enzyme, results in an inhibition of chlorophyll b degradation, and hence a stay-green phenotype (Sato et al. 2009).
More than 180 color mutants were hitherto isolated and characterized in rice, and at least 80 genes (Deng et al. 2014; Sun et al. 2017). These mutations distributed in 12 chromosomes were cloned, most of which are controlled by a pair of recessive nuclear genes (Qian et al. 1996; Wang et al. 2008; Xie et al. 1995). In this study, we characterized a white lesion mimic leaves1 (wlml1) mutant, which was derived from an M2 population of the indica rice cultivar Taichung Native 1 (TN1) after ethyl methanesulfonate (EMS) mutagenesis. This mutant displays white and lesioned leaves, and the mutant is stably inherited, segregating as a recessive mutation in a nuclear gene. We characterized the mutant phenotype, including pigment contents, chloroplast structure, and plant responses to different temperatures, and further delimited the WLML1 locus to a 159.7-kb region on chromosome 4. The fine mapping of this gene lays the foundation for the cloning of related genes and leaf color breeding programs, as well as research on chlorophyll metabolism and mechanisms of disease resistance in plants.
Materials and methods
Plant materials and growth conditions
The wlml1 mutant was derived from the M2 population of the indica rice cultivar TN1 after EMS mutagenesis. The wlml1 mutant and its progeny all exhibited white and diseased leaves, and this phenotype was stably inherited over multiple generations. The wlml1 mutant and the japonica cultivar Wuyunjing 7 (W7) were used to make the segregating population for mapping. All plants were grown in the paddy fields at the China National Rice Research Institute (CNRRI) in Hangzhou (summer) and Lingshui (winter), China.
Agronomic trait measurements
Wild-type TN1 and wlml1 plants were planted in the same paddy fields at the CNRRI in Hangzhou in the summer of 2017, and the same growth periods were maintained for all plants. The main agronomic traits (plant height, tiller, leaf length, leaf width, spike length, number of primary branches, and 1000-grain weight) were investigated once the plants reached maturity. Data from three biological replicates were compiled for each trait.
Photosynthetic rate measurement
In the morning, a photosynthetic apparatus (Li-6800, Li-Cor, Lincoln, Nebraska, USA) was used to measure the photosynthetic rate of both the white and diseased parts of corresponding sites on leaves from mutant and wild-type plants that had reached maturity. Three biological replicates were used.
Observing temperature sensitivity in seedlings
Mature seeds from wlml1 and TN1 plants were soaked in sterile water in a 96-well plate in the same growth environment for 15 days. Seedlings that had grown to a similar size were transferred to a growth chamber (Panasonic, MLR-352H-PC) for 7 days under 12 h light/12 h dark at constant temperatures of either 20, 28, or 40 °C. To measure the thermo-sensitivity of the plants, phenotypes were observed, chlorophyll pigment contents were measured, and the expression levels of thermo-sensitivity related genes were quantified.
Pigment content measurement
During the mature stage, corresponding sections were cut from the leaves of TN1 and wlml1 plants, immersed in extraction solution (mixture of 2/3 95% acetone and 1/3 ethanol) and incubated in the dark at 26 °C for 24 h. The optical density of sample solutions was measured with an ultraviolet spectrophotometer (UV-2600) at 663, 645, and 470 nm. Three biological replicates were conducted for each plant type. The contents of chlorophyll (Chl a and Chl b) and carotenoid (Car) in each leaf sample was calculated according to Arnon’s method (Arnon 1949).
Observation of chloroplast ultrastructure
At the four-leaf stage, leaves from wild-type and wlml1 mutant plants were cut into small pieces, fixed with 2.5% glutaraldehyde (diluted with phosphate buffer, pH 7.2), and placed under vacuum until the specimens were immersed. For mutant plants, both green and white sections of the leaves were collected for comparison. Specimens were then fixed, dehydrated, embedded, sliced, dyed, and then examined and photographed using a transmission electron microscope (Hitachi H-7650, Tokyo, Japan) (Li et al. 2010; Wu et al. 2016).
Genetic analysis and fine mapping
To identify the gene responsible for the wlml1 phenotype, we first examined its segregation. To that end, we performed a reciprocal cross between wlml1 and the indica cultivar TN1. The phenotypes of plants in the F1 and F2 populations were observed and analyzed using the χ2 test.
For fine mapping, an F2 segregating population was derived from the crosses between the wlml1 mutant and the japonica cultivar W7. The parents and 2878 F2 individuals were planted in the paddy field. Among the F2 individuals, 645 that displayed the mutant phenotype were used to map the WLML1 gene. 23 wlml1/W7 F2 individuals with the mutant phenotype were used for linkage analysis. All F2 plants displaying the leaf color mutant phenotype were used for fine mapping. Total genomic DNA was extracted from fresh leaves using the CTAB (cetyltrimethylammonium bromide-based) method.
Insertion/deletion (InDel) and simple sequence repeat (SSR) markers were developed based on the sequence differences between the two rice varieties, japonica Nipponbare (http://rgp.dna.affrc.go.jp) and indica 93-11 (http://www.rise.genomics.org.cn). The 232 SSR markers were uniformly distributed among 12 chromosomes and the primer sequences for the molecular markers are listed in Supplemental Table 1.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Real-time fluorogenic quantitative PCR was performed to analyze leaf color-related gene expressions in the wild type and the wlml1 mutant. The total RNAs in the wild type and the mutant leaves at the four-leaf stage were extracted to synthesize the first-strand cDNA by reverse transcription. The volume of the PCR system was 10 µL and contained the following: 1 µL of cDNA template, 0.5 µL of 10 µM each primer (F/R), 5 µL of 2 × SYBR (TOYOBO), and ddH2O up to 10 µL. The PCR conditions were: 1 min at 95 °C, followed by 45 cycles of 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 1 min. Real-time PCR was performed with a Real-Time PCR System (ABI 7900). The expression levels were normalized against the expression of Histone H3. The primers used for the quantification of related genes are listed in Supplemental Table 2.
Statistical analysis
All results are expressed as mean values (± standard deviation, SD) based on three biological replicates. Statistical significance was assessed using Student’s unpaired t test. Probability values of < 5% were considered to be statistically significant, single asterisk (*) and double asterisk (**) represents the significant at the level of 5 and 1%, respectively. The statistical analysis of the gene relative expression levels, pigment content, photosynthetic rate and agronomic traits in this study all follow the methods described above.
Results
Phenotypic and agronomic traits analysis
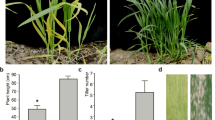
At the four-leaf stage, wlml1 mutant leaves displayed yellow patches and some lesions that were not present on the leaves of wild-type (TN1) plants (Fig. 1a–c). These yellow patches changed to white, and the lesions increased in abundance on the white patches (Fig. 1c–e). At the mature growth stage, the agronomic traits (effective tiller number, panicle length, total grain number per panicle, grain length, grain width, and seed-setting rate) were not significantly different between the wild-type and mutant plants; however, plant height and grain weight were significantly reduced in the mutant (Table 1).
Phenotype comparison between wild-type TN1 and wlml1 mutant plants at various growth stages. a–c Seedlings and leaves from wild-type TN1 and wlml1 mutant plants at the four-leaf stage. d–f Pictures of seedlings and leaves from wild-type TN1 and wlml1 mutant plants at the mature stage. Scale bars represent 2 cm (a, e, f); 1 cm (b, c); and 10 cm (d)
Analysis of photosynthetic rate, photosynthetic pigment content, and chloroplast ultrastructure
The leaf-color phenotype observed in the same wlml1 plant was not consistently exhibited in all leaves (variegated phenotype), so the photosynthetic rate and photosynthetic pigments were measured by sub-area. Compared to the wild-type rates (Fig. 2a), the photosynthetic rates of the white sections and the lesioned sections of mutant leaves declined by 34 and 36.3%, respectively. The content of Chl a, Chl b, and carotenoid (Car) pigments in the white leaf sections from the wlml1 mutant declined by 56.6, 56.5, 55.6%, respectively (Fig. 2c), but the pigment contents did not differ between green leaf sections from the wlml1 mutant and those from wild-type plants (Fig. 2b).
Photosynthetic rate, photosynthetic pigment content, and chloroplast ultrastructure of wild-type TN1 and wlml1 mutant plants. a Photosynthetic rates of the white sections and lesioned sections of wlml1 mutant leaves compared with corresponding sections from wild-type TN1 leaves. b Content of photosynthetic pigments in green leaves from wild-type TN1 and wlml1 mutant plants. c Content of photosynthetic pigments in white leaves from wlml1 mutants compared with leaves from wild-type TN1 plants. d–k Chloroplast ultrastructure in leaves from wild-type TN1 plants (d, h), and green (e, i), white (f, j), and lesioned (g, k) sections of leaves from wlml1 mutant plants. FW fresh weight, Ch chloroplast, N nucleus, G grana stacks, A amyloplast, OG osmophilic plastoglobuli. Data represent the mean ± SD of three biological replicates (Student’s t test: *P < 0.05, **P < 0.01)
To explore the cause of the observed decline in photosynthetic pigments in wlml1 plants, we observed difference of chloroplast ultrastructures in the leaves from TN1 and wlml1 plants at the four-leaf stage by transmission electron microscopy. The chloroplasts in the green sections of leaves from both TN1 (Fig. 2c, g) and wlml1 (Fig. 2d, h) plants were all intact. However, the white-section chloroplasts of wlml1 leaves were severely degraded, there were few normal chloroplast structures; In addition, the number of chloroplasts decreased and an absence of the grana lamellae (Fig. 2e, i).
Genetic analysis and fine mapping of WLML1
A genetic analysis was conducted on the F1 and F2 populations that were generated from reciprocal crosses between wlml1 mutants and wild-type TN1 plants. The leaf color of all F1 plants from either cross (wlml1 × TN1 or TN1 × wlml1) was consistent with that of the wild type. Out of the 656 F2 plants from the wlml1 × TN1 cross, 496 displayed the wild-type phenotype and 160 displayed the mutant phenotype. Out of the 524 F2 plants from the TN1 × wlml1 cross, 389 displayed the wild-type phenotype and 135 displayed the mutant phenotype. In both crosses, the segregation ratio of the wild-type to the mutant phenotype was approximately 3:1 according to a χ2 test (Table 2). The genetic analysis showed that WLML1 was a single recessive nuclear gene.
The F2 population generated from the wlml1 × W7 cross was used for fine mapping of WLML1. WLML1 was initially mapped between markers P1 and P2 on chromosome 4 according to 23 individuals displaying the wlml1 phenotype and 232 pairs of SSR markers (Fig. 3a). To further define WLML1, the mapping population was expanded to 645 plants and new SSR and InDel markers were designed between P1 and P2 according to sequence differences between the rice varieties japonica Nipponbare (http://rgp.dna.affrc.go.jp) and indica 93-11 (http://www.rise.genomics.org.cn). Finally, the locus was delimited to a 159.7-kb region between markers P15 and P16. According to the genomic annotation database RAP-DB (http://rapdb.dna.affrc.go.jp/), this region contains 30 predicted Open Reading Frames (ORFs) (Fig. 3b–d).
Fine mapping of WLML1. aWLML1 was initial mapped to a region on chromosome 4 between markers P1 and P2. b Fine mapping of WLML1 using 645 F2 plants from the wlml1 × W7 cross. The numerals indicate the number of recombinants. c WLML1 was located between markers P15 and P16 within a 159.7-kb genomic DNA region. d 30 ORFs are candidates for WLML1
Relative expression of genes related to leaf color in leaves from wild-type and mutant plants
The leaves from wlml1 mutant plants display white patches and lesions. Changes in leaf color are associated with chlorophyll metabolism and chloroplast development. Therefore, we examined the transcript abundance of genes associated with chlorophyll synthesis, photosynthesis, chlorophyll degradation, and chloroplast development in wlml1 mutant and wild-type seedlings. We found that genes associated with chlorophyll synthesis (DVR, CHLH, OsPORA, OsPORB, and CAO1) were significantly down-regulated, in wlml1 mutants compared with wild-type plants, while most chlorophyll degradation genes (SGR and OsPAO) were significantly up-regulated, except for OsPCCR (Fig. 4a). Some genes related to chloroplast development were down-regulated (PsbA, PsaB, V2, V3, LhcP2 and Rps15), while others were up-regulated (V1, RpoC1, and RpoC2) (Fig. 4b).
Relative expression of genes related to leaf color in leaves from wild-type TN1 and wlml1 mutant plants. a Relative expression of genes associated with chlorophyll synthesis and degradation between wild-type TN1 and wlml1 mutant plants. b Relative expression of genes associated with chloroplast development between wild-type TN1 and wlml1 mutant plants. c Relative expression of genes associated with disease resistance and ROS between wild-type TN1 and wlml1 mutant plants
The formation of lesions in leaves is also related to disease resistance and production of reactive oxygen species (ROS). All tested disease resistance-related genes (PBZ1, PR1A, PR1B, and PR10) and antioxidant related genes (OsCATB, OsAPX2, and OsPOD1) were significantly up-regulated (Fig. 4c).
The wlml1 mutant is temperature-sensitive
To identify whether the phenotype in the wlml1 mutant was affected by temperature at the seedling stage, we transplanted 15-day-old wlml1 and TN1 seedlings into growth chambers at different temperatures (20, 28 and 40 °C) for 7 days. There were no obvious differences in leaves color between TN1 and wlml1 plants at 40 °C, although green withered leaf tips were observed (Fig. 5a, d), and Chl a and Chl b contents were increased in wlml1, compared with TN1 plants (Fig. 5g). At 28 °C, leaves were slightly yellow (Fig. 5b, e), and Chl a and Car contents were significantly reduced in wlml1 plants, compared to wild type plants (Fig. 5h). At 20 °C, the leaves of mutant plants were also yellow and displayed a decrease in pigment content compared to the wild-type plants, but this difference was more obvious than that at 28 °C (Fig. 5c, i). Furthermore, the lesions appeared in leaves from the wlml1 mutant plants at 20 °C but not evident in wild-type plants (Fig. 5f).
Leaf phenotypes and photosynthetic pigment contents of wild-type and wlml1 seedlings grown at 40 °C (a, d, g), 28 °C (b, e, h), and 20 °C (c, f, i). Data represent the mean ± SD of three biological replicates (Student’s t test: *P < 0.05, **P < 0.01). FW fresh weight. Scale bars are 2 cm (a–c); and 1 cm (d–f)
As the temperature decreased, genes associated with chlorophyll degradation (NYC3, NOL, OsPCCR, OsPAO, and SGR), chloroplast development (V1, V2, and Cab1) and oxidation resistance (OsCATA, OsCATB, and OsPOD1) were significant up-regulated in wlml1 compared with TN1 (Fig. 6). For example, the ratio of expression of these genes NYC3, OsPCCR, V1, V2, OsCATA, and OsPOD1 between wlml1 and TN1 were 0.88, 0.93, 0.73, 1.09, 0.98, and 0.98 at 40 °C; and 0.96, 1.2, 0.96, 1.1, 1.09, and 1.42 at 28 °C; and 2.6, 3.15, 4.48, 3.09, 3.1, and 3.53 at 20 °C, respectively.
Relative expression of genes related to chlorophyll degradation, chloroplast development and oxidation resistance under different temperatures in wlml1 and wild-type seedlings. a Relative expression of genes related to chlorophyll degradation, chloroplast development, and oxidation resistance at 40 °C (a), 28 °C (b) and 20 °C (c). Data represent the mean ± SD of three biological replicates (Student’s t test: *P < 0.05, **P < 0.01)
Discussion
Leaf color markers are widely used in maintaining the genetic purity of seeds and as a marker of self-pollination to monitor male sterile lines (Deng et al. 2014; Lv et al. 2013). The only phenotypic difference between wild type and the wlml1 mutant is a slight difference in plant height and 1000-grain weight (Table 1). Therefore, wlml1 can be used as a visible marker in rice breeding.
There are seven genes related to leaf color on chromosome 4 that have been cloned in rice: Rice Lesion Initiation 1 (OsRLIN1), Mitochondrial PPR25 (OsMPR25), Albino Midrib 1(OsAM1), ENT-Kaurene Synthase-Like 2 (OsKS2), Young Seedling Stripe1 (OsYSS1), Giant Chloroplast (OsGIC), and Wax Crystal-sparse Leaf 1 (OsWSL1). RLIN1 encodes a putative coproporphyrinogen III oxidase from the tetrapyrrole biosynthesis pathway, and the rlin1 mutant showed a lesion mimic phenotype (Wang et al. 2015; Sun et al. 2011). MPR25 encodes a pentatricopeptide repeat protein that belongs to the E subgroup of the pentatricopeptide repeat protein family. The mpr25 mutant exhibits growth retardation and pale-green leaves with reduced chlorophyll content during the early stages of plant development (Toda et al. 2012). AM1 encodes a putative potassium efflux antiporter, and the am1 mutant exhibited a green/white variegation phenotype in the first few leaves, followed by an albino midrib phenotype in older tissues (Sheng et al. 2014; Nonomura et al. 2011). OsKS2 encodes a functional ent-beyerene synthase that is responsible for the biosynthesis of various diterpenoids (Tezuka et al. 2015), and the mutant has short, dark green leaves (Ji et al. 2014). YSS1 encodes a chloroplast nucleoid-associated protein that is essential for chloroplast development in rice seedlings. The yss1 mutant develops striated leaves at the seedling stage, but no abnormal phenotype in leaf 5 and onwards (Zhou et al. 2017). GIC encodes a PARC6 homolog, and the gic mutant has fewer and enlarged chloroplasts (Kamau et al. 2015). WSL1 encodes a multiple organellar RNA editing factor, and wsp1 mutants exhibit green and white sectors starting from the three-leaf stage and not entirely green panicles at the heading stage (Zhang et al. 2017). In this study, the WLML1 locus was mapped to a 159.7-kb DNA region on chromosome 4. Both the location of the WLML1 and the mutant phenotype are different from the known leaf color mutants, so we speculate that WLML1 is a new gene.
Together, the characteristics of the wlml1 mutant phenotype of fewer chloroplasts, severe chloroplast degradation, and decreased pigment content (Fig. 2), as well as the up- or down-regulation of genes associated with chlorophyll metabolism and chloroplast development suggest that chloroplast dysfunction causes the leaf color change in the wlml1 mutant (Fig. 4a, b).
ROS function as signals for the activation of defense genes. ROS also drive the cross-linking of cell wall structural proteins, function as a local trigger of programmed death in pathogen-challenged cells, and as a diffusible signal for the induction of genes encoding cellular protectants in adjacent cells (Bradley et al. 1992; Lamb and Dixon 1997; Montillet et al. 2005; Levine et al. 1994). The appearance of a lesion mimic phenotype (Fig. 1), and the up-regulation of disease resistance and ROS-related genes (Fig. 4c) in wlml1 compared with wild-type plants suggests that the wlml1 phenotype may be related to an imbalance of ROS.
The observed changes in pigment contents of the wlml1 mutant when grown at various temperatures (Fig. 5h–j) implies that WLML1 function may not be completely lost in the wlml1 mutant, and was regulated by temperature. At 20 °C, the ratio of expression of genes associated with chlorophyll degradation and chloroplast development are up-regulated between wlml1 and TN1 compared to at 28 and 40 °C. This suggests that WLML1 regulates leaf color by accelerating chlorophyll degradation, and chloroplast development-associated genes, possibly by feedback regulation. Furthermore, the formation of lesions and higher expression of antioxidant-related genes in wlml1 mutants grown at 20 °C compared to 28 and 40 °C suggests that WLML1 may participate in regulating biotic and abiotic stress in rice (Fig. 6).
Conclusion
In this study, we identified the wlml1 mutant, which has white and lesioned leaves and can be used as a marker in breeding to maintain the purity of varieties due to the minimal impact of the mutation on important agronomic traits in rice. We mapped WLML1 to a 159.7-kb region on chromosome 4, which laid the foundation for subsequent map-based cloning. Since the wlml1 mutation negatively affected chlorophyll metabolism and chloroplast development, this mutant will be useful for in-depth studies of chlorophyll metabolism and chloroplast development. Since the mutant phenotype was influenced by temperature, this mutant can also be used to study regulation of chloroplast development at different temperatures. In addition, the wlml1 mutant has a lesion mimic phenotype that is affected by temperature. Therefore, we speculate that the gene encoded at WLML1 could be related to resistance to disease and stress, which we plan to investigate in future experiments. If this speculation is verified, wlml1 will be useful for research on plant disease and stress resistance.
References
Arnon DI (1949) Copper enzymes in isolated chloroplasts, polyphenoxides in Beta vulgaris. Plant Physiol 24:1–15
Bradley DJ, Kjellbom P, Lamb CJ (1992) Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell 70:21–30
Deng XJ, Zhang HQ, Wang Y, He F, Liu JL, Xiao X, Shu ZF, Li W, Wang GH, Wang GL (2014) Mapped clone and functional analysis of leaf-color gene ygl7 in a rice hybrid (Oryza sativa l. ssp. indica). PLoS ONE 9:e99564
Fromme P, Melkozernov A, Jordan P, Krauss N (2003) Structure and function of photosystem i: interaction with its soluble electron carriers and external antenna systems. FEBS Lett 555:40–44
Ge CW, E ZG, Pan JJ, Jiang H, Zhang XQ, Zeng DL, Dong GJ, Hu J, Xue DW (2015) Map-based cloning of a spotted-leaf mutant gene ossl5, in japonica, rice. Plant Growth Regul 75:595–603
Hu Z, Peng LP, Cai YH (1981) A yellow-green nucleus mutant of rice. Acta Genet Sin 8:256–261 (Chinese)
Ji SH, Gururani MA, Lee JW, Ahn BO, Chun SC (2014) Isolation and characterisation of a dwarf rice mutant exhibiting defective gibberellins biosynthesis. Plant Biol 16:428–439
Kamau PK, Sano S, Takami T, Matsushima R, Maekawa M, Sakamoto W (2015) A mutation in giant chloroplast encoding a parc6 homolog affects spikelet fertility in rice. Plant Cell Physiol 56:977–991
Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48:251
Levine A, Tenhaken R, Dixon R, Lamb C (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79:583–593
Li C, Li DZ, Dong YJ, Ye SH, Zhang XM (2010) Genetic analysis and mapping of a thermo-sensitive white stripe leaf mutant at the seedling stage in rice (Oryza sativa). Chin J Rice Sci 24:223–227
Li X, He Y, Yang J, Jia YH, Zeng HL (2018) Gene mapping and transcriptome profiling of a practical photo-thermo-sensitive rice male sterile line with seedling-specific green-revertible albino leaf. Plant Sci Int J Exp Plant Biol 266:37
Lin BY, Yu HJ (1995) Inheritance of a striped-leaf mutant is associated with the cytoplasmic genome in maize. Theor Appl Genet 91:915–920
Liu CL, Wei XJ, Shao GN, Chen ML, Hu PS, Tang SQ (2012) Research progress of the molecular mechanism of leaf color mutation in rice. China Rice 18:15–21
Lv Q, Xu J, Wu P (2013) Ospapst1, a useful mutant for identifying seed purity and authenticity in hybrid rice. Plant Signal Behav 8:2036–2048
Marco GD, D’Ambrosio N, Giardi MT, Massacci A, Tricoli D (1989) Photosynthetic properties of leaves of a yellow green mutant of wheat compared to its wild type. Photosynth Res 21:117–122
Montillet JL, Chamnongpol S, Rustérucci C, Dat J, Van De Cotte B, Agnel JP Battesti C, Inzé D, Van BF, Triantaphylidès C (2005) Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol 138:1516–1526
Nagata N, Satoh S, Tanaka R, Tanaka A (2004) Domain structures of chlorophyllide a oxygenase of green plants and prochlorothrix hollandica in relation to catalytic functions. Planta 218:1019–1025
Nonomura KI, Eiguchi M, Nakano M, Takashima K, Komeda N, Fukuchi S, Miyazaki S, Miyao A, Hirochika H, Kurata N (2011) A novel RNA-recognition-motif protein is required for premeiotic G1/s-phase transition in rice (Oryza sativa L.). Plos Genetics 7:e1001265
Palmer RG, Rodriguez de Cianzio S (1985) Conditional lethality involving nuclear and cytoplasmic chlorophyll mutants in soybeans. Theor Appl Genet 70:349
Qian Q, Zhu XD, Zeng DL, Zhang XH, Yan XQ, Xiong ZM (1996) Study on the new special material white-green seedlings controlled by cytoplasmic gene. China Seeds 4:11–12 (Chinese with English abstract)
Rudoi AB, Shcherbakov RA (1998) Analysis of the chlorophyll biosynthetic system in a chlorophyll b-less barley mutant. Photosynth Res 58:71–80
Sato Y, Morita R, Katsuma S, Nishimura M, Tanaka A, Kusaba M (2009) Two short-chain dehydrogenase/reductases, non-yellow coloring 1 and nyc1-like, are required for chlorophyll b, and light-harvesting complex II degradation during senescence in rice. Plant J 57:120–131
Sheng P, Tan J, Jin M, Wu F, Zhou K, Ma W, Heng Y, Wang J, Guo X, Zhang X, Cheng Z, Liu L, Wang C, Liu X, Wan J (2014) Albino midrib 1, encoding a putative potassium efflux antiporter, affects chloroplast development and drought tolerance in rice. Plant Cell Rep 33:1581–1594
Su Y, Hu S, Zhang B, Ye W, Niu Y. Guo L, Qian Q (2017) Characterization and fine mapping of a new early leaf senescence mutant es3(t), in rice. Plant Growth Regul 81(3):419–431
Sun C, Liu L, Tang J, Lin A, Zhang F, Fang J, Zhang G, Chu C (2011) RLIN1, encoding a putative coproporphyrinogen III oxidase, is involved in lesion initiation in rice. J Genet Genom 38:29–37
Sun LT, Lin TZ, Wang YL, Ni M, Hu TT, Liu SJ, Wan J (2017) Phenotypic analysis and gene mapping of a white stripe mutant st13 in rice. Chin J Rice Sci 31(4):355–363
Tanaka A, Tanaka R (2006) Chlorophyll metabolism. Curr Opin Plant Biol 9:248–255
Tezuka D, Ito A, Mitsuhashi W, Toyomasu T, Imai R (2015) The rice ent-kaurene synthase like 2 encodes a functional ent-beyerene synthase. Biochem Biophys Res Commun 460:766
Toda T, Fujii S, Noguchi K, Kazama T, Toriyama K (2012) Rice mpr25 encodes a pentatricopeptide repeat protein and is essential for RNA editing of nad5 transcripts in mitochondria. Plant J 72:450
Wang F, Wang G, Li X, Huang J, Zheng J (2008) Heredity, physiology and mapping of a chlorophyll content gene of rice (Oryza sativa L.). J Plant Physiol 165:324
Wang P, Gao J, Wan C, Zhang F, Xu Z, Huang X, Sun X, Deng X (2010) Divinyl chlorophyll(ide) a can be converted to monovinyl chlorophyll(ide) a by a divinyl reductase in rice. Plant Physiol 153:994–1003
Wang J, Ye B, Yin J, Yuan C, Zhou X, Li W, He M, Wang J, Chen W, Qin P, Ma B, Wang Y, Li S, Chen X (2015) Characterization and fine mapping of a light-dependent leaf lesion mimic mutant 1 in rice. Plant Physiol Biochem 97:44
Wang ZW, Lv J, Xie SZ, Zhang Y, Qiu ZN, Chen P, Cui YT, Niu YF, Hu SK, Jiang HZ (2018) Ossla4, encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling growth in rice. Plant Growth Regul 84:249–260
Wu LW, Ren DY, Hu SK, Li GM, Dong GJ, Jiang L, Hu XM, Ye WJ, Cui YT, Zhu L, Hu J, Zhang GH, Gao ZY, Zeng DL, Qian Q, Guo LB (2016) Mutation of OsNaPRT1 in the NAD salvage pathway leads to withered leaf tips in rice. Plant Physiol 171:1085–1098
Xie R, Zhu FY, Deng XH, Zuo YS, Yang ZL, Wu LJ (1995) A preliminary study on the temperature sensitive chlorophyll natural mutant in dual-purpose genic male sterile rice. Southwest China J Agric Sci 8:124–128
Yang Y, Xu J, Huang L, Leng Y, Dai L, Rao Y. Chen L, Wang Y, Tu Z, Hu J, Ren D, Zhang G, Zhu L, Guo L, Qian Q, Zeng D (2016) Pgl, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J Exp Bot 67:1297
Ye WJ, Hu S, Wu L, Ge C, Cui Y, Chen P, Xu J, Dong G, Gu L, Qian Q (2017) Fine mapping a major QTL qfcc7-1, for chlorophyll content in rice (Oryza sativa L.) cv. PA64s. Plant Growth Regul 81:81–90
Zhang L, Gao YM (2009) Advance in gene mapping and cloning of leaf color mutants in rice. Crop 2:12–16
Zhang Z, Cui X, Wang Y, Wu J, Gu X, Lu T (2017) The RNA editing factor WSP1 is essential for chloroplast development in rice. Mol Plant 10:86–98
Zhou K, Ren Y, Zhou F, Wang Y, Zhang L, Lyu J, Wang Y, Zhao S, Ma W, Zhang H, Wang L, Wang C, Wu F, Zhang X, Guo X, Cheng Z, Wang J, Lei C, Jiang L, Li Z, Wan J (2017) Young seedling stripe1, encodes a chloroplast nucleoid-associated protein required for chloroplast development in rice seedlings. Planta 245:1–16
Acknowledgements
This study was supported by the Natural Science Foundation of China (Grant Nos. 31461143014, 31771778), the National Key Research and Development Program of China (Grant No. 2016YFD0100902-07).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no potential competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Chen, P., Hu, H., Zhang, Y. et al. Genetic analysis and fine-mapping of a new rice mutant, white and lesion mimic leaf1. Plant Growth Regul 85, 425–435 (2018). https://doi.org/10.1007/s10725-018-0403-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-018-0403-7