Summary
This study evaluated the preclinical activity and molecular predictors of response to gefitinib (Iressa®, Astra Zeneca Inc, UK) in nasopharyngeal carcinoma (NPC). The activity of gefitinib was evaluated in four human NPC cell lines—HK1, HONE-1, CNE2, C666-1. A representative gefitinib-sensitive (HK1, IC50 = 250 nM) and gefitinib-resistant cell line (HONE-1, IC50 > 15 μM) were selected and compared for expression of epidermal growth factor receptor (EGFR) and related ligands, and activation of downstream proteins. Gefitinib induced G1 cycle arrest, apoptosis and inhibited cell invasion more significantly in HK1 than HONE-1 cells. HK1 expressed higher levels of p-EGFR, lower p-AKT and phospho-signal transducer and activator of transcription 3 (p-STAT3) than other cell lines. EGFR gene was found to be amplified in HK1. Gefitinib at IC50 concentrations significantly suppressed EGF-induced activation of p-EGFR, phospho-mitogen-activated protein kinase (p-MAPK) and p-STAT3, but p-AKT showed persistent activation in HK1 and HONE-1 cells. There was no difference in EGFR-ligand expression between the 4 NPC cell lines. In NPC samples derived from non-responders to gefitinib, 50% and 60% showed cytoplasmic and nuclear pi-EGFR expression, respectively, and 33% showed p-AKT expression. EGFR or KRAS mutations were not detected. This study suggests that most NPC cell lines are intrinsically resistant to gefitinib (except HK1 cells), and further studies are needed to confirm whether EGFR gene amplification and persistent AKT activation may influence response to gefitinib in NPC.
Similar content being viewed by others
Introduction
Nasopharyngeal carcinoma (NPC) is endemic to China and the Southeast Asian region and areas like Hong Kong, where it reaches a peak incidence rate of around 20 per 100,000 person-years [1]. Our previous work supports epidermal growth factor receptor (EGFR) as a potential therapeutic target for NPC, as it is overexpressed in 80% of NPC and represents a negative prognostic factor [2]. Based on our work which has demonstrated the preclinical and clinical activity of the anti-EGFR antibody, cetuximab in NPC [3, 4] in NPC, we evaluated the EGFR tyrosine kinase inhibitor, gefitinib (ZD1839, Iressa®, Astra Zeneca Inc, Macclesfield, UK) in a previously reported phase II trial in advanced NPC [5]. The objectives of the current study were to determine the preclinical activity and molecular predictors of response to gefitinib in NPC cell lines and in primary tumor biopsies obtained from our phase II study. These factors included the activation level of EGFR and its downstream signaling mediators (mitogen-activated protein kinase, MAPK; protein kinase B, AKT; signal transducer and activator of transcription 3, STAT3); expression level of EGFR-related ligands (epidermal growth factor, EGF; amphiregulin; heparin-binding EGF-like growth factor, HB-EGF; transforming growth factor alpha, TGF-α); and the presence of any EGFR or KRAS mutations.
Method and materials
Cell culture
Analyses were carried in four human-derived NPC cell lines (CNE-2, C666-1, HONE-1 and HK1) and the normal nasopharyngeal epithelial cell line NP69 was used as a control.
Reagents and antibodies
Gefitinib was kindly provided by Astra Zeneca Inc (Macclesfield, UK). For in vitro experiments, the following antibodies were purchased from Cell Signaling Technology (Beverly, USA): anti-total EGFR, anti-phospho(p)-EGFR (Y1068), anti-total STAT3, anti-p-STAT3 (Tyr705); anti-total p44/42MAPK(Thr202/Tyr204), anti-p-p44/42MAPK(Thr202/Tyr204), AKT (#9272S), anti-total AKT and p-AKT (Ser473) (#9270S), antibody against cleaved poly(ADP-ribose) polymerase (PARP) proteins. Anti-beta-actin antibody was from Calbiochem, Merck (Gibbstown, USA). The following antibodies from Cell Signaling Technology (Beverly, USA) were used for immunohistochemistry: p-AKT (Ser473), anti-p-EGFR (Tyr 845), and p-EGFR (Tyr 992). MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was from Invitrogen (USA) and recombinant human EGF was purchased from PeproTech (London, UK).
Cell viability assay
Cells were treated with increasing concentrations of gefitinib (from 0 to 100 μM) for up to 48 hours (h) and cytotoxicity was assessed by the MTT assay. Cells were cultured in 48-well plates (2,500–7,000 cells/well) and then incubated with gefitinib at 24 h for 48 to 72 h. DMSO was used as a blanking control. The drug concentration at which 50% of maximum growth inhibition was achieved was determined as the IC50 using the GraphPad PRISM software (ISS, San Diego, CA). All experiments were repeated in triplicates.
Western blot and apoptosis
Western Blot was used to detect the expression of total and phosphorylated levels of EGFR, MAPK, STAT3 and AKT in NPC cell lines, at basal condition and after treatment with gefitinib at or near IC50 concentration for 48 h under serum-starved and EGF-stimulated (20 ng/ml) conditions. Apoptosis was determined via detection of PARP cleavage proteins. Cell lysates were prepared as previously described [6] and 25 µg of total protein was subjected to SDS-PAGE and immunoblotting with specific antibodies as listed above. Supersignal West Pico Chemiluminescence Western blotting detection reagents (ThermoFisher Scientific, USA) were used for signal detection by autoradiography.
RNA extraction and RT-PCR
The expression levels of EGFR-related ligands were determined by Reverse Transcription-Polymerase Chain Reaction (RT)-PCR. Total RNA was extracted from cell pellets using TriReagent (Molecular Research Center, USA) and treated with DNase by a TURBO DNA-free™ Kit (Ambion, USA). One μg of RNA was used for reverse transcription using the Go-Taq polymerase (Promega Co, USA) using GAPDH as control as previously described [7]. The primers and conditions are outlined in Table A of the section on Supplementary Material. RT-PCR products were analyzed on a 1.8% agarose gel.
Cell cycle
Cells were plated in 100-mm2 Petri dishes (3 to 4 × 105) and then treated with gefitinib for up to 24 h. Cells were collected by trypsinization, fixed with 70% cold ethanol and stored at −20°C. DNA staining was performed with a solution containing RNase (0.2 mg/ml) and propidium iodide (0.05 μg/ml). Analysis was performed using a FACScan flow cytometer and data were processed with CELLQuest software (Becton Dickinson Co, USA).
Matrigel invasion assay
The cell invasion assay was performed using a Matrigel Invasion Chamber with a pore size of 8 µm (Becton Dickinson Co, USA). Cells were seeded at a density of 25 × 104 cells per well (for HONE-1 cells), or 7 × 104 cells per well (for HK1 cells) in serum-free medium. The cell-containing chamber was immersed in a lower chamber containing either serum-free medium with or without gefitinib (5 μM, 15 μM), in the presence of EGF (10 ng/ml) or DMSO. After 24 h, non-invaded cells retained in the upper chamber were removed and stained with 1% Toludine Blue O (Sigma, USA) in 1% Borax (USB, USA), and then counted under the microscope at 200 times magnification.
EGFR gene amplification
DNA copy number changes in NPC cell lines were analyzed by Agilent Human Genome CGH Microarray and scanned with a G2505B Agilent DNA microarray scanner as previously described [8]. Using the CGH Analytics Software 3.4, the array CGH data was analyzed and genomic profile of each sample was generated. All software and equipment used for microarray were from Aglient Technologies (USA). Z-score algorithm was applied for calculating the chromosomal aberration, and genes with log2 ratio greater than 1 were considered as amplified.
Immunohistochemistry
Paraffin-embedded samples of primary NPC biopsies were obtained before treatment from 9 participants (out of a total of 15) of a phase II trial of gefitinib in NPC [5]. This clinical protocol was approved by the Institutional Ethics Committee and all participants gave written informed consent. Tissue sections of 4 μm thickness were immunostained using the Benchmark XT immunostainer and ultraView™ detection kit (Ventana Inc, USA). An immunostained sample of breast cancer was used as a positive control and the negative control was made by replacing the antibody with Tris buffered saline. Slides were evaluated under a light microscope at 200 times magnification by a pathologist (K.F.To). Phosphorylated-AKT (Ser473) expression was considered positive (1+) or negative, if nuclear staining was present or negative, respectively. The expression of pi-EGFR-Y845 was scored by estimating the proportion of tumor cells with positive nuclear staining (negative if none; 1+, if less than or equal to 10%; 2+, if 10–50%; 3+, if over 50%). The cytoplasmic expression of pi-EGFR-Tyr 992 was scored as previously described [2].
Mutational analysis
Paraffin-embedded NPC samples were deparafinized and DNA was isolated using QIAamp DNA Mini Kit (Qiagen GmbH, Germany). Sections of 130 μm thick were cut and dewaxed through suspension in 40 µL of TK buffer. To inactivate the enzyme, samples were incubated at 55°C for 3 days and heated at 95°C for 10 min. One µL of protein was used for PCR analysis, the amplicons were amplified using AmpliTaq Gold polymerase (Applied Biosystems, USA) and purified using the High Pure PCR Purification Kit (Roche Diagnostics, USA). Sequencing was performed using the dRhodamine Terminator Cycle Sequencing Kit (Perkin-Elmer, USA) and the Applied Biosystems 310 Genetic Analyzer. The primers and PCR conditions are outlined in Table B of the section on Supplementary Material.
Results
Effect of gefitinib on growth, apoptosis, cell cycle progression and cell invasion
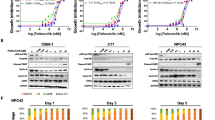
The IC50 value for gefitinib in HK1 cells (250 nM) was the lowest compared with the other NPC cell lines of which the IC50 values ranged between 15–40 µM, while the IC50 for NP69 was 28.6 μM (data was not shown). In order to elucidate the biological characteristics associated with sensitivity to gefitinib, HK1 and HONE-1 cell lines were chosen as a representative gefitinib-sensitive and gefitinib-resistant cell line, respectively, for all subsequent analyses described below.
When treated with gefitinib at IC50 concentrations, apoptosis was observed in both HK1 and HONE-1 cells after 24 to 48 h as evident by an increased expression of cleaved PARP (Fig. 1a and b). Exposure to gefitinib at similar concentrations also resulted in G1 cycle arrest (Fig. 1c), and inhibition of the invasive activity of HONE-1 and HK1 cells compared with control (Fig. 2).
a Effect of gefitinib (Iressa®, 15 μM) on apoptosis in HONE-1 cells as indicated by expression of cleaved PARP. b Effect of gefitinib (Iressa®, 250 nM) on apoptosis in HK1 cells as indicated. c Gefitinib induced G1 Arrest in HONE-1 (left) and HK1 (right) cell lines following exposure for 24 h at IC50 concentrations by expression of cleaved PARP
Molecular profile of gefitinib-sensitive and gefitinib insensitive cell lines
At basal condition, the level of total and p-EGFR and p-MAPK was higher in HK1 compared to HONE-1 cells (Fig. 3a). In contrast, the level of p-AKT and p-STAT3 were higher in HONE-1 compared with HK1 cells, while there were no significant differences in the levels of total AKT, STAT3 and MAPK expression. However, such differences were not observed under EGF-stimulated conditions (Fig. 3b). When HK1 and HONE-2 cells were treated with gefitinib at or near their respective IC50 concentrations, the level of p-MAPK and p-STAT3 were significantly reduced in both cell lines (Fig. 3c). Phosphorylated AKT expression was significantly reduced when both cell lines were treated with gefitinib well above the respective IC50 concentrations (Fig. 3b). The expression level of TGF-α, EGF, HB-EGF, amphiregulin were compared in HK1 and HONE-1 cell lines. No difference was found in the ligand expression level of these cell lines (Fig. 3d). Furthermore, none of the cell lines harbored EGFR TK mutations (data not shown). In order to explain why HK1 cells expressed such high level of p-EGFR in the absence of EGFR activating mutations, HK1 cells were analyzed for gene amplification and a high level of EGFR amplification was found using aCGH microarray platform (Fig. 4). The threshold log2 ratio of this amplicon core was around 3.
a Basal expression of activated and total EGFR and its downstream mediators in NPC cell lines (serum-starved condition). b Expression of downstream mediators of EGFR in HK1 and HONE-1 at basal, serum-starved, EGF-stimulated (20 ng/ml) and above-IC50 concentration of gefitinib (Iressa®). c Expression of downstream mediators of EGFR in HK1 (250 nM) and HONE-1 (15 μM) at or near IC50 concentration of gefitinib. d Expression of EGFR ligands in NPC cell lines
Immunohistochemistry and EGFR mutation in tumor samples
Of the pre-treatment biopsies obtained from 9 patients, only 3 samples showed mild (+1) p-AKT expression. Under half (4/9 samples) of the samples showed EGFR overexpression (cytoplasmic staining), while the 6 (out of 9) samples showed mild to moderate nuclear expression of EGFR (see Table 1 and Fig. 5). None of the tumors analyzed harbor KRAS or EGFR mutations (data not shown). Since clinical response was not observed in the previously reported phase II study [5], correlation between immunohistochemical results and clinical response was not performed.
Immunohistochemical staining for phosphorylated (p-) EGFR and p-AKT-ser473 in NPC samples. a & d. Immunohistochemical, IHC. staining for EGFR-Tyr845. a Representative image showing strong nuclear (grade 3) positivity (case 10, original magnification, Mag. × 200). d A case with negative staining (case 3, original mag. × 200). b & e IHC staining for EGFR-Tyr992. b Representative image showing strong cytoplasmic (grade 3) positivity (case 9, original Mag. × 200). e A case with negative staining (case 3, original mag. × 200). c & f IHC staining for pAKT-ser473. c Representative image showing grade 1 positivity (case 9, original mag. × 200). f A case with negative staining (case 5, original mag. × 200)
Discussion
In this study, gefitinib was found to inhibit cell growth and invasion, induce apoptosis and G1 cycle arrest in human NPC cell lines, as consistent with previous reports in other cancer types [9, 10]. However, the relatively high concentrations required to achieve these effects suggested that most NPC cell lines are intrinsically resistant to gefitinib, with the exception of HK1. Furthermore, treatment with gefitinib at IC50 concentrations significantly inhibited EGF-induced phosphorylation of important downstream mediators of EGFR, namely, STAT3 and MAPK to a similar extent in both HK1 and HONE-1 cells, but AKT activation appeared to remain persistent in both cell lines at similar concentration. These results prompted the need to elucidate the molecular basis of these changes by evaluating the activation level of EGFR and its downstream signaling mediators, ligand expression, presence of EGFR mutations and gene amplifications in NPC, by comparing these molecular profiles between a representative gefitinib-sensitive (HK1) and a resistant cell line (HONE-1).
The 2 cell lines differed in the basal activation level of EGFR and downstream mediators, in that HK1 cells expressed a higher level of p-EGFR and p-MAPK, while HONE-1 expressed a higher level of p-AKT and p-STAT3. However, MAPK, AKT and STAT3 activation were similar in both cell lines under EGF-stimulated condition, suggesting that these signaling axes are inherently intact. Previous studies in other cancer types have associated resistance to gefitinib with increased level of MAPK (e.g. prostate cancer cells) [11] and AKT (e.g. in lung [12] and glioma cells [13]. As consistent with our study, increased STAT3 activation can be found in gefitinib-resistant glioma cells, despite the fact that STAT3 could be easily inhibited at low doses of gefitinib [13]. As in our NPC cell lines, higher concentrations of gefitinib was required to inhibit AKT and MAPK activation compared with EGFR in gefitinib-resistant glioma cells, suggesting that the AKT signaling maybe an escape mechanism for the inhibitory effect of gefitinib [13].
EGFR was found to be amplified in the gefitinib-sensitive HK1 cell lines. Gain in gene copy number or EGFR gene amplification is a common event in NPC, and further validation is needed to elucidate whether EGFR amplification may contribute to the enhanced sensitivity towards gefitinib in NPC [14]. This is supported by studies in lung cancer samples, where increased copy number of EGFR has been associated with better response to gefitinib [15].
Increased expression of EGFR-related ligands have been shown to predict resistance to EGFR TK inhibitors in vitro, these include overexpression of TGF-α in breast and pancreatic cancer cells [16, 17], and amphiregulin in breast and lung cancer cells [16, 18]. In gefitinib-resistant breast cancer cells, exposure to gefitinib led to increased nucleo-cytoplasmic trafficking of EGFR ligands, thereby suggesting the presence of an intracrine feedback loop [16]. However, the current study did not identify any differences in expression of EGFR ligands between HK1 and HONE-1 cell lines, thus excluding the possibility that reduced basal activation of STAT3 and AKT in HK1 cells might be related to altered autocrine signaling in NPC.
Our in vitro data suggests that resistance to gefitinib may be associated with lower level of EGFR activation and persistent AKT activation in NPC cell lines, therefore we examined tumor samples obtained from gefitinib-resistant participants of a previously reported phase II study [5]. The study was terminated after 15 patients were entered because no clinical response was observed, hence the small number of biopsies obtained for the current study. Around 33% (3/ 9 samples) of tumors weakly expressed p-AKT, which concurred with the reported 40% p-AKT overexpresssion rate in previous reports [19, 20]. Over 50% (5/9 samples) of the current cohort showed no cytoplasmic EGFR expression, while over 60% expressed moderate to strong nuclear EGFR expression—the latter which is a negative prognostic factor in some cancers [21]. This was unexpected as our previous report found that over 80% of NPC overexpressed cytoplasmic EGFR [2]. However, the small number of tumor samples available makes it difficult to draw conclusions. The low level of AKT activation in NPC tumors from gefitinib-resistant patients is consistent with previous studies in lung cancer, where the lack of p-AKT expression was associated with poorer clinical outcome following gefitinib [12, 22].
In summary, a high level of basal EGFR activation (possibly from EGFR gene amplification) is an important indicator for sensitivity to gefitinib, while further studies are needed to confirm whether persistent AKT activation maybe associated with resistance. Tumoral status of EGFR activation and gene amplification should be further evaluated as biomarkers for selecting patients in clinical trials involving EGFR inhibitors.
References
Tao Q, Chan AT (2007) Nasopharyngeal carcinoma: molecular pathogenesis and therapeutic developments. Expert Rev Mol Med 9:1–24
Ma B, Poon T, To KF, Teo PML, Chan ML, Mo KF, Ma B, Lai MP, Ho S, Chan ATC (2003) Expression and prognostic significance of epidermal growth factor receptor and HER2 protein in nasopharyngeal carcinoma. Head Neck 25:864–872
Sung FL, Poon TC, Hui EP, Ma BB, Liong E, To KF, Huang DP, Chan AT (2005) Antitumor effect and enhancement of cytotoxic drug activity by cetuximab in nasopharyngeal carcinoma cells. In Vivo 19:237–245
Chan AT, Hsu MM, Goh BC, Hui EP, Liu TW, Millward MJ, Hong RL, Whang-Peng J, Ma BB, To KF, Mueser M, Amellal N, Lin X, Chang AY (2005) Multicenter, phase II study of cetuximab in combination with carboplatin in patients with recurrent or metastatic nasopharyngeal carcinoma. J Clin Oncol 23:3568–3576
Ma B, Hui EP, King A, To KF, Mo F, Leung SF, Kam M, Lo YM, Zee B, Mok T, Ahuja A, Chan AT (2008) A phase II study of gefitinib patients with metastatic or locoregionally recurrent nasopharyngeal carcinoma and evaluation of plasma Epstein-Barr virus DNA as a biomarker of efficacy. Cancer Chemother Pharmacol 62:59–64
Lui VW, Boehm AL, Koppikar P, Leeman RJ, Johnson D, Ogagan M, Childs E, Freilino M, Grandis JR (2007) Antiproliferative mechanisms of a transcription factor decoy targeting signal transducer and activator of transcription (STAT) 3: the role of STAT1. Mol Pharmacol 71:1435–1443
Seng TJ, Low JS, Li H, Cui Y, Goh HK, Wong ML, Srivastava G, Sidransky D, Califano J, Steenbergen RD, Rha SY, Tan J, Hsieh WS, Ambinder RF, Lin X, Chan AT, Tao Q (2007) The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene 26:934–944
Adelaide J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F, Charafe-Jauffret E, Cervera N, Desplans J, Parzy D, Schoenmakers E, Viens P, Jacquemier J, Birnbaum D, Bertucci F, Chaffanet M (2007) Integrated profiling of basal and luminal breast cancers. Cancer Res 67:11565–11575
Hopfner M, Sutter AP, Huether A, Schuppan D, Zeitz M, Scherubl H (2004) Targeting the epidermal growth factor receptor by gefitinib for treatment of hepatocellular carcinoma. J Hepatol 41:1008–1016
Bonaccorsi L, Carloni V, Muratori M, Formigli L, Zecchi S, Forti G, Baldi E (2004) EGF receptor (EGFR) signaling promoting invasion is disrupted in androgen-sensitive prostate cancer cells by an interaction between EGFR and androgen receptor (AR). Int J Cancer 112:78–86
Festuccia C, Gravina GL, Millimaggi D, Muzi P, Speca S, Ricevuto E, Vicentini C, Bologna M (2007) Uncoupling of the epidermal growth factor receptor from downstream signal transduction molecules guides the acquired resistance to gefitinib in prostate cancer cells. Oncol Rep 18:503–511
Han SW, Hwang PG, Chung DH, Kim DW, Im SA, Kim YT, Kim TY, Heo DS, Bang YJ, Kim NK (2005) Epidermal growth factor receptor (EGFR) downstream molecules as response predictive markers for gefitinib (Iressa, ZD1839) in chemotherapy-resistant non-small cell lung cancer. Int J Cancer 113:109–115
Li B, Chang CM, Yuan M, McKenna WG, Shu HK (2003) Resistance to small molecule inhibitors of epidermal growth factor receptor in malignant gliomas. Cancer Res 63:7443–7450
Hui AB, Lo KW, Teo PM, To KF, Huang DP (2002) Genome wide detection of oncogene amplifications in nasopharyngeal carcinoma by array based comparative genomic hybridization. Int J Oncol 20:467–473
Takano T, Ohe Y, Sakamoto H, Tsuta K, Matsuno Y, Tateishi U, Yamamoto S, Nokihara H, Yamamoto N, Sekine I, Kunitoh H, Shibata T, Sakiyama T, Yoshida T, Tamura T (2005) Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol 23:6829–6837
Ferrer-Soler L, Vazquez-Martin A, Brunet J, Menendez JA, De Llorens R, Colomer R (2007) An update of the mechanisms of resistance to EGFR-tyrosine kinase inhibitors in breast cancer: Gefitinib (Iressa)-induced changes in the expression and nucleo-cytoplasmic trafficking of HER-ligands (Review). Int J Mol Med 20:3–10
Pino MS, Shrader M, Baker CH, Cognetti F, Xiong HQ, Abbruzzese JL, McConkey DJ (2006) Transforming growth factor alpha expression drives constitutive epidermal growth factor receptor pathway activation and sensitivity to gefitinib (Iressa) in human pancreatic cancer cell lines. Cancer Res 66:3802–3812
Kakiuchi S, Daigo Y, Ishikawa N, Furukawa C, Tsunoda T, Yano S, Nakagawa K, Tsuruo T, Kohno N, Fukuoka M, Sone S, Nakamura Y (2004) Prediction of sensitivity of advanced non-small cell lung cancers to gefitinib (Iressa, ZD1839). Hum Mol Genet 13:3029–3043
Yip WK, Leong VC, Abdullah MA, Yusoff S, Seow HF (2008) Overexpression of phospho-Akt correlates with phosphorylation of EGF receptor, FKHR and BAD in nasopharyngeal carcinoma. Oncol Rep 19:319–328
GM MJA, Pathmanathan R, Raab-Traub N (2004) Differential signaling pathways are activated in the Epstein-Barr virus-associated malignancies nasopharyngeal carcinoma and Hodgkin lymphoma. Cancer Res 64:5251–5260
MK LSY, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC (2001) Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol 3:802–808
Cappuzzo F, Magrini E, Ceresoli GL, Bartolini S, Rossi E, Ludovini V, Gregorc V, Ligorio C, Cancellieri A, Damiani S, Spreafico A, Paties CT, Lombardo L, Calandri C, Bellezza G, Tonato M, Crino L (2004) Akt phosphorylation and gefitinib efficacy in patients with advanced non-small-cell lung cancer. J Natl Cancer Inst 96:1133–1141
Acknowledgements
This study was supported by the Research Grant Council Competitive Earmarked Research Grant (Reference 4445/05M). Result of this study was presented in part at the Annual Meeting of the American Association of Clinical Oncology 2007, abstract 3171, April 14–18, 2007, Los Angeles, CA. We thank Astra Zeneca Inc for providing gefitinib in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Brigette Ma & Vivian Lui are joint 1st authors.
An erratum to this article can be found at http://dx.doi.org/10.1007/s10637-010-9397-3
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1
(DOC 44 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Ma, B.B., Lui, V.W., Poon, F.F. et al. Preclinical activity of gefitinib in non-keratinizing nasopharyngeal carcinoma cell lines and biomarkers of response. Invest New Drugs 28, 326–333 (2010). https://doi.org/10.1007/s10637-009-9316-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10637-009-9316-7