Abstract
Historical hunting pressures on koalas in the southern part of their range in Australia have led to a marked genetic bottleneck when compared with their northern counterparts. There are a range of suspected genetic disorders such as testicular abnormalities, oxalate nephrosis and microcephaly reported at higher prevalence in these genetically restricted southern animals. This paper reports analysis of differential expression of genes from RNAseq of lymph nodes, SNPs present in genes and the fixation index (population differentiation due to genetic structure) of these SNPs from two populations, one in south east Queensland, representative of the northern genotype and one in the Mount Lofty Ranges South Australia, representative of the southern genotype. SNPs that differ between these two populations were significantly enriched in genes associated with brain diseases. Genes which were differentially expressed between the two populations included many associated with brain development or disease, and in addition a number associated with testicular development, including the androgen receptor. Finally, one of the 8 genes both differentially expressed and with a statistical difference in SNP frequency between populations was SLC26A6 (solute carrier family 26 member 6), an anion transporter that was upregulated in SA koalas and is associated with oxalate transport and calcium oxalate uroliths in humans. Together the differences in SNPs and gene expression described in this paper suggest an underlying genetic basis for several disorders commonly seen in southern Australian koalas, supporting the need for further research into the genetic basis of these conditions, and highlighting that genetic selection in managed populations may need to be considered in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Koalas (Phascolarctos cinereus) are an iconic Australian marsupial species listed as vulnerable on the IUCN ‘red list’ of threatened species (Woinarski and Burbidge 2016). While a large part of their ongoing population decline is due to habitat loss, disease threats are additionally limiting population viability (Polkinghorne et al. 2013). Following European settlement, koala populations across Australia declined significantly due to hunting in the 1890s to 1920s, with southern populations nearing extinction. During this time, small refuge populations were established on offshore Victorian islands and these koalas have been subsequently used to restock most of their former southern range. These southern populations (Victoria and South Australia (SA)) are genetically distinct from the northern animals (Queensland (QLD) and New South Wales) (Neaves et al. 2016; Ruiz-Rodriguez et al. 2016). Southern koalas are in over-abundance in some parts of their range and they have also recently been shown to have lower rates of the two major disease threats affecting koala population viability, chlamydiosis and koala retrovirus (KoRV) (Simmons et al. 2012; Polkinghorne et al. 2013; Legione et al. 2016; Fabijan et al. 2019a, 2019b; Sarker et al. 2019).
Koalas in the southern areas have restricted genetic diversity when compared with their northern counterparts (Houlden et al. 1996, 1999; Taylor et al. 1997; Fowler et al. 1998; Lau et al. 2014; Ruiz-Rodriguez et al. 2016; Kjeldsen et al. 2018) likely as an effect of the founder, bottleneck and inbreeding effects in these populations. Low diversity places populations at risk of increased expression of recessive deleterious traits. A number of pathologies with a suspected genetic basis have been reported in the southern populations at a higher rate than in the northern animals. Testicular abnormalities have been recorded in animals derived from the Kangaroo Island population in SA including testicular aplasia (Seymour et al. 2001; Cristescu et al. 2009), asymmetry and atrophy (Speight et al. 2018) with rates as high as 12.9% of affected males reported in the Eyre Peninsula population in SA. In contrast testicular aplasia was found to be absent in the outbred Pilliga ranges in New South Wales, with the rates of testicular abnormalities closely linked to the inbreeding co-efficients of the populations (Seymour et al. 2001).
Another suspected genetic disorder is the renal disease oxalate nephrosis, which has a very high prevalence in the Mount Lofty Ranges population in SA, with up to 32% of deaths attributed to this cause (Speight et al. 2018). Koalas with this disease have kidney failure associated with deposition of calcium oxalate crystals in the tubules of the kidneys. It is also apparent that even clinically normal animals in the Mount Lofty and Kangaroo Island populations have elevated levels of oxalate in their urine when compared with an unaffected northern population from southeast QLD (Speight et al. 2013, 2014). Genetic causes of renal oxalate disease are reported in other species (Danpure and Rumsby 2004; Heiene et al. 2009; Vidgren et al. 2012) but these are yet to be identified in southern koalas.
Microcephaly, colloquially known as “pin-head” has also been reported in southern animals, particularly by wildlife care groups working with certain isolated populations in Victoria (Australian Senate 2011), though with no formal scientific studies of the problem it is hard to gauge the prevalence of this condition. Other skeletal disorders reported across the range of koalas that may have a genetic component include craniofacial tumours (primarily osteochondromas) (Sutton 1986; Canfield et al. 1990; Hanger and Loader 2014), microphthalmia and scoliosis (Higgins et al. 2018).
Approaches to analysing the genetic diversity of bottlenecked wildlife populations have generally focussed on examining a limited range of markers. Probably the most popular technique has been the use of variability in short tandem repeats (STR) also known as microsatellite markers via PCR and visualisation or sequencing of variations in the number and size of these repeats across individuals and populations. PCR amplification and sequencing of variable gene regions such as whole or partial mitochondrial genomes or genes or major histocompatibility (MHC) loci has also been extensively deployed. The advent of the availability of reference genomes has also led to the use of SNP analysis, typically performed using a microarray from a related species for which extensive genetic information on SNP variability is available (Hindrikson et al. 2017; O'Brien et al. 2017; Gienapp et al. 2017; Cammen et al. 2016). With newer sequencing technologies becoming more affordable partial genome sequencing technologies such as RADseq (restriction site associated DNA sequencing) (Andrews et al. 2016; Cammen et al. 2016) have also been deployed. The lack of good quality reference sequences and the costs of whole genome sequencing have meant that more comprehensive methods such as whole genome resequencing and GWAS as deployed in humans and some high value domestic species has remained limited (Jones and Good 2016). The numbers of animals required for meaningful analysis of population variance in association with phenotypic traits has remained prohibitive for wildlife species. However studies in North American puma have demonstrated that for species with significant genetic bottlenecks and large tracts of heterozygosity in their genomes, whole genome sequencing of individuals can provide startlingly in depth information on inbred populations (Saremi et al. 2019). The use of RNAseq (exome capture) in human and livestock species studies of genetic disorders is a widely used compromise in terms of ability to provide in-depth SNP sequencing for larger numbers of individuals for a reasonable cost (in comparison to whole or partial genome sequencing) (Jones and Good 2016). Its use in wildlife species has however remained limited, presumably because of the increased difficulty in preservation and sample quality from field sample collection sites as well as the lack of reference genomes to facilitate mapping of sequencing reads (Cammen et al. 2016).
This paper reports a novel use of RNAseq data to identify the likely underlying genetic variation responsible for a number of suspect genetic disorders in a bottlenecked wildlife population (southern koalas). The paper reports the identification of high quality variants in transcriptomic data and differential gene expression of genes in lymph node tissue from a total of 29 koalas from two populations, one from south east QLD (10 animals), representative of the “northern” genotype and one from the Mount Lofty Ranges in SA (19 animals) representative of the “southern” genotype. This paper provides the first evidence for an underlying genetic basis for several disorders affecting southern koala populations and sets the stage for more detailed studies to inform breeding, translocation and restocking decisions in this iconic vulnerable species.
Methods
Ethics
Ethical approval for this study was granted by the University of Queensland Animal Ethics Committee, permit number ANFRA/SVS/461/12 and ANRFA/SVS/445/15, the Queensland Government Department of Environment and Heritage Protection permit number WISP11989112, the University of Adelaide Animal Ethics Committee permit number S-2013-198 and the South Australian Government Department of Environment, Water and Natural Resources Scientific Research Permit Y26054.
Samples
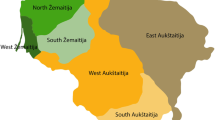
Samples were collected from wild-rescued koalas that had been euthanised for clinical reasons and submitted for post-mortem examinations from South East QLD (Greater Brisbane) (n = 10) and SA (Mount Lofty Ranges) (n = 19) Fig. 1. The mainland Mount Lofty Ranges koala population in SA primarily originated from the Kangaroo Island population (derived in turn from Victorian island populations) but also included koalas from QLD and New South Wales (Lindsay 1950; Bergin 1978). Age was determined by dentition and the amount of wear on the upper premolar (Martin and Handasyde 1999). Submandibular lymph nodes were collected within 2–6 h of death into RNALater® and stored at − 80 °C.
Map of koala sampling locations, Mt Lofty Ranges marked with a blue star, Greater Brisbane with a green star. Approximate current range of wild koalas marked with a blue line (current distribution derived from Atlas of living Australia http://www.ala.org.au accessed 22/12/20) estimated demarcation between southern and northern genotypes marked with a green line
Details of the koalas, sample quality and read quantity are provided in Online resource 1. A summary of koala details is provided in Table 1. Details of the comparative pathology, chlamydial and Koala retrovirus (KoRV) status of the animals included in this study are reported elsewhere (Tarlinton et al. 2017; Sarker et al. 2019; Fabijan et al. 2020).
RNA extraction and sequencing
Total RNA was extracted using an RNeasy Mini kit with on column DNAase1 digestion (Qiagen). RNA quantity and quality were assessed via anXpose spectrophotometer (Bioke) and Agilent 2100 Bioanalyzer. mRNA was prepared for sequencing using the Illumina TruSeq stranded mRNA library prep kit and 100 base pair, paired end sequencing was performed on an Illumina HiSeq. Reads were trimmed with skewer (Jiang et al. 2014) using 3′ quality cut off of 3 (− q = 3) and mean quality of 20 (− Q = 20) prior to mapping to the available koala genome contig assembly GCF_002099425.1_phaCin_unsw_v4.1 from NCBI genomes for SNP detection or gene expression analysis.
Genome mapping and SNP calling
Trimmed reads were mapped to the koala contigs using Hisat2 (Kim et al. 2019). Sorted bam files were processed using opossum (Oikkonen and Lise 2017) to allow robust identification of SNPs from transcriptome data. SNPs were called from combined bam files using samtools (Li et al. 2009)mpileup and output genotype likelihoods generated in bcf format (-g -b -u options). Bcftools (BCFtools)was used to call multiallelic SNPs (-vm options). Identified SNPs were filtered using vcftools (Danecek et al. 2011). SNPs where a base was missing in more than half the samples, with minor allele frequency less than 0.2, reads of minimum quality < 30 were removed (--max-missing 0.5 --maf 0.2 --minQ 30 --recode). For each SNP a population FST was calculated using Weir and Cockerham’s algorithm (Wier and Cockerham 1984) (--weir-fst-pop option). Locations and predicted effects of SNPs on the function of known genes were determined using SNPEff (Cingolani et al. 2012) with maximum up/down stream distance set as 2000 bp.
Gene expression analysis
To quantitate the expression of known genes, sequence alignment and read quantification was performed using the pseudo-alignment-based tool Kallisto v0.43.0 (Bray et al. 2016). Transcripts with a mean TPM < 1 for both groups (QLD and SA) were removed. Differential expression of transcripts between QLD and SA animals was determined using Sleuth v0.28.1 (Pimentel et al. 2017). Transcripts with a false detection rate corrected p value < 0.05 and a mean log2 fold change (log2FC) greater than 1 and less than minus 1 were considered to be differentially expressed. False detection rate correction was performed using the Benjamini–Hochberg method (Benjamini and Hochberg 1995). To account for possible confounding factors analysis was repeated with a model comparing location alone against one containing location + age (as approximated by tooth wear class) + sex. This is by a likelihood ratio test in sleuth of a “full model” (~ location + age + sex) vs a”reduced model” (~ location).
Protein structural changes
Protein structural changes due to SNPs in the gene SLC26A6 were predicted using the Phyre2 software suite (Kelley et al. 2015).
Gene set enrichment analysis was conducted using a hypergeometric test in ToppGene. (Chen et al. 2009) Benjamini–Hochberg corrected p values < 0.05 are reported.
Results
Identification of SNP variants between populations
A total of 215,834 unique variants were identified. To remove SNPs with high coverage variability, those where a base was identified in ≤ 66% of both QLD and SA populations were removed. This resulted in a final set of 135,191 SNPs. The mean fixation index (FST) of these SNPs was 0.412; when filtered for those SNPs with strongest evidence of divergence between the two populations (FST ≥ 0.8), 24,981 variants remained (23,145 SNPs and 1836 INDELs, see Online resource 2). A high FST is evidence of a history of separation of genes or populations but could be biased by variants on the sex chromosomes. Therefore, it was important to test if any of the high FST variants were biased by sex of the animals. As the genome of the koala is currently not assembled into chromosomes, but rather as contigs, separation of variants that are located on the sex chromosomes was not immediately possible. As a proxy to identify any variants associated with sex of samples, a Wilcoxon rank sum test was used to test the frequency of each different genotype in male vs female animals. Only a single variant (contig NW_018344261.1 position 1080527) showed a significant association with sex (p value = 0.048). This however was not significant after correcting for multiple hypothesis testing. Therefore, we propose that the SNPs reported are not biased by the ratio of sexes of the animals in the two populations.
Annotation of the variants using SNPEff highlighted the majority of variants are located in introns (34,488 variants). As the mapped reads are from RNAseq, possible sources of high coverage in introns are a mix of mis-annotation, immature mRNA sequencing or possible genomic DNA contamination. To keep analysis here as robust as possible, these predicted intronic variants were not used in later analysis. The majority of other annotations were in non-coding regions (3′ UTR = 10,256, downstream = 7658, upstream = 2504, 5′UTR = 1700). Of the SNPs in coding regions, the majority were synonymous variants (5589) and a smaller proportion were non-synonymous variants (3320).
Functional annotation of population variants
SNPEff characterises the predicted impact of SNP on gene function. This annotation ranges from unknown “modifier” to low, moderate and high impact. Removing those genes with limited impact (“modifier”) resulted in 3669 unique variant positions affecting 2168 unique genes (Online resource 2). Gene set enrichment showed significant enrichment in poor brain function in humans (DisGeNET (Piñero et al. 2020) codes C3714756, C0917816, C0423903, C4020876, C1843367, C0025362 respectively) and Human Phenotypes relating to brain abnormalities (Human Phenotype Ontologies (Köhler et al. 2019) codes 0002060, 0100547, 0011282, 0011283 respectively). A total of 349 genes were associated with these terms which contained a total of 2341 variants (Online resource 3 and 4).
Differential gene expression between populations
A comparison of gene expression between the QLD and SA koalas identified 104 statistically significant differentially expressed transcripts in the lymph node tissue (Online resource 5). Forty of these were downregulated in SA animals (compared with QLD animals) and 64 of these were upregulated in SA animals (compared with QLD animals). As samples were recovered from wild rescue euthanised animals and couldn’t be perfectly matched between locations there may be confounding factors which contribute to gene expression changes observed. Factors that could be more confidently controlled for were sex and age. However, age is an approximation based on tooth wear class (see online resource 1) and so is used with caution. Using the same tools but with a model for differential expression analysis controlling for age and sex, 51 statistically significant differentially expressed transcripts were detected (q value < 0.05, 26 upregulated in SA, 25 downregulated). These are highlighted in online resource 5 in column “qval (~ location + age + sex)”.
There were a variety of transcripts downregulated in the SA animals that have a plausible link to testicular abnormalities, including several with testes specific expression or a role in sex steroid metabolism (at least in the mouse and human models) including Meiotic Kinetochore factor (MEIKIN), serine protease inhibitor Kazal-type 10-like (important in sperm capacitance), lamin tail domain containing 1 (LMNTD1), multiple EGF like domains 11 (MEGF11), growth arrest specific 8 (GAS8), aldo–keto reductase family 1 member C1 homolog and critically, the androgen receptor (AR). There were also a number of transcripts that are usually testis- expressed that were upregulated in the SA animals including ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 (ST6GALNAC2), family with sequence similarity to 204 member A (FAM204A), anaphase promoting complex subunit 16 (ANAPC16), testis expressed 11 (TEX11), zinc finger protein 345-like (testes expressed transcription factor), nicotinamide nucleotide adenylyltransferase 3 (NMNAT3) and PHD finger protein 24-like. However, none of the SA koalas in the study cohort exhibited testicular abnormalities.
In terms of anion-exchange transport proteins known to be linked to oxalate metabolism disturbances (and therefore to oxalate nephrosis), solute carrier family 26 member 6 (SLC26A6) was upregulated in the lymph node of SA animals, of which 21% (4/19) were affected by clinical oxalate nephrosis. The functions of this protein (in humans) include apical membrane anion-exchange, a role in the pH buffering system and mediation of intestinal chloride absorption and oxalate secretion.
Similar to the genes enriched in the SNP analysis there were a large number of differentially expressed genes between the southern and northern koala groups that are linked to known brain expression and facial deformity syndromes in people. Including calcium dependent secretion activator (CADPS), protocadherin gamma-C4, family with sequence similarity 204 member A (FAM204A), vesicle amine transport 1 like (VAT1L), alkaline ceramidase 3 (ACER3), nestin (NES), ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 (ACAP3), dishevelled binding antagonist of beta catenin 1 (DACT1), neuronal PAS domain protein 1 (NPAS1), prosaposin-like, acyl-CoA oxidase 2 (ACOX2). None of the SA koalas in the study cohort exhibited brain or facial deformities, but two koalas showed scoliosis and kyphosis.
There were a small number of genes differentially expressed that could plausibly be linked to the overwhelming KoRV expression in the QLD koalas and lower prevalence in SA koalas. These genes include a C–C motif chemokine 4-like transcript and integrin alpha-M-like, prostaglandin E synthase (PTGES) which were upregulated in the SA animals. C–C motif chemokine 4 binds directly to CCR5 (one of the receptors for HIV in humans) and inhibits CCR5-mediated entry of HIV-1 and HIV replication. The solute carrier family 29 member 1 (Augustine blood group) (SLC29A1), which was downregulated in SA animals, is thought to be a DC-sign-like pathogen-associated molecular pattern (PAMP) that is potentially involved in viral recognition. Similarly, down regulation of several MHC1 linked transcripts (HLA class I histocompatibility antigen, alpha chain G-like, class I histocompatibility antigen, Gogo-OKO alpha chain-like), which could be involved in KoRV recognition, were seen in the SA animals.
Several of the differentially expressed genes appear to be transcript variants of the same locus, in some cases different transcripts of the same gene are upregulated or downregulated in the SA animals, these include Subtilisin-like protease one (2 variants, one upregulated, one downregulated), SUN domain-containing protein 2-like (4 variants, 3 upregulated, 1 down regulated), bridging integrator 2 (BIN2) (1 up regulated, 1 down regulated), pyroglutamyl-peptidase I (PGPEP1) (1 upregulated, 1 down regulated) and syncytin-1- like mRNA (2 up regulated and 1 down regulated).
SNPs associated with differentially expressed genes
A total of 241 variants were detected within 52 transcripts from 47 unique differentially expressed genes (see online resource 6). A permutation test to determine the joint probability any transcript is differentially expressed and also contains a SNP showed that this is unlikely to have occurred by chance alone (p < 1 × 10−16, calculated from probability of 10,000 sample permutations). Of these the majority (116) are located in annotated introns or downstream of genes (38 downstream and 35 3′ UTR). A smaller proportion of variants were upstream (7 variants) and 5′ UTR located (4 variants). A total of 40 variants were located in coding regions (21 synonymous, 14 missense and other splice variants). A Kruskal–Wallis rank sum test was used to test for significant differences of within gene expression between observed genotypes at each variant positions (that is, if different SNP genotypes are associated with different levels of cis gene expression). A total of 80 variant positions showed significantly different gene expression between genotypes (Benjamini–Hochberg corrected p value < 0.05). These are within 8 genes: CUTA, LETMD1, CD209 antigen-like protein E (LOC110202118), class I histocompatibility antigen (LOC110203128), SLC26A6, SLC29A1, ST6GALNAC2 and NPAS1. As with all variants studied, the majority (48) were in introns with 24 in the coding sequences (17 synonymous and 7 missense) and the majority appear to be fixed within the SA animals compared with the QLD animals. Three genes had SNPs in the 3′ UTR; LETMD1 (3 SNPs); LOC110202118 and ST6GALNAC2 (each with one SNP each) which may be related to gene expression changes through mRNA degradation or miRNA binding.
The functional relationship of some of these genes to the disorders in the southern populations is not clear (CUTA, LETMD1, SLC29A1, ST6GALNAC2) and the population variation seen in these SNPs may be the result of genetic divergence of the two geographically separated populations, through genetic drift or due to the genetic bottlenecks that the SA population has undergone. Two of these genes (LOC110202118: CD209 antigen-like protein E, LOC110203128: class I histocompatibility antigen) were linked to immune function and may be related to either expression in the face of KoRV infection or differing resistance to KoRV infection (as has been reported for some rare MHC alleles in HIV). NPAS1 is expressed in neuronal tissue and may be linked to the plethora of other neuronal genes differentially expressed between populations, potentially explaining reports of facial and neural deformities in some southern populations.
The SLC26A6 variants that were differentially expressed are alternatively spliced transcripts of this gene at the 5′ end (Fig. 2). There were two nucleic acid changes compared with the reference sequence present. The first was a T to C at position 11 in transcript XM_020998995.1, resulting in a leucine to proline change in the amino acid sequence at this location (and in the corresponding location in transcript XM_020999000.1). This could result in a shortening of the initial alpha helix of this protein’s secondary structure (Online resource 7). The second was a G to C change at position 1924 in XM_020998995.1 and the corresponding location in all other differentially expressed alternatively spliced transcripts of this gene resulting in a conservative valine to leucine change that nonetheless could also result in a shortening of an alpha helix in the protein secondary structure (Online resource 7). In both cases the QLD animals were either homozygous or heterozygous for the version present in the reference genome while the SA animals were uniformly homozygous for the alternate SNP (Fig. 3) displaying a significantly higher expression. The four animals from SA with confirmed pathology consistent with oxalate nephrosis (samples SAR1352A10, SAR2433A1, SAR2433A5 and SAR2433A6) show higher than average expression of transcript XM_020999000.1 compared to other SA animals (Fig. 4).
Percentage of animals with each combination of SNPs in SLC26A6 transcripts (./. stands for missing in that transcript. Line 1 = G to C at position 1924 (G1924C) of XM_020998995.1, line 2 = T to C at position 11 (T11C) of XM_020998995.1. Orange = QLD animals, Blue = SA animals. QLD animals are homo- or heterozygous for reference nucleotide. The SA animals were uniformly homozygous for alternate nucleotides
Transcript expression levels of the missense variants of transcript XM_020998995.1 at position 11 in SLC26A6. SA animals (triangles) with the G genotype (C in mRNA) have a higher expression level (TPM normalised expression) than QLD animals (circles) with either the A (T in mRNA) or G (C in mRNA) genotype. The four animals from SA with confirmed pathology consistent with oxalate nephrosis (samples SAR1352A10, SAR2433A1, SAR2433A5 and SAR2433A6) are highlighted as red triangles
Discussion
This paper takes a novel approach in the use of RNAseq data from two geographically distinct koala populations (one of which is genetically restricted) to examine differentially expressed genes and differentially segregated genes that are potentially linked to disease syndromes seen in the genetically restricted population. While such approaches are used in human and domesticated or semi-domesticated species to explore specific genetic traits (Cammen et al. 2016; Steiner et al. 2013; Jones and Good 2016), their application to wildlife species has not been previously developed.
The most striking link between the data in this study and disease in southern koalas is that of the anion transporter SLC26A6 which transports anions, including oxalate, in the kidney and intestine. This gene is both differentially expressed and demonstrates differential segregation of SNPs between the populations. This anion transporter has been the focus of several recent studies in mice and rat models, where knockout animals with this gene functionally removed demonstrated a propensity to the development of hyperoxaluria and/or oxalate urolithiasis (Freel et al. 2006; Jiang et al. 2006). Conversely, overexpression of renal SLC26A6 has also been found to lead to increased oxalate excretion and renal oxalate deposition in rats (Jiang et al. 2018), showing the complexity of the mechanisms of oxalate secretion. Case control studies in humans with oxalate urolithiasis are also exploring the role of SLC26A6 in oxalate uroliths in people, so far demonstrating a link between high expression of the gene (whether functional or not is not known) and oxalate kidney stones in people (Monico et al. 2008; Lu et al. 2016).
Based on our findings it is likely that SLC26A6 is either directly or indirectly linked to the high prevalence of oxalate nephrosis and hyperoxaluria seen in the SA animals. The SNPs in the SLC26A6 gene may be fixed in this population, and in the animals tested in this study this appears so. However only four of the 19 koalas (21%) in the study showed clinical oxalate nephrosis, suggesting that more complex factors may be involved in disease manifestation. It is tempting to speculate that the SNPs identified here affect the protein tertiary structure and reduce its effectiveness in oxalate secretion, driving a compensatory (but ineffective) feedback loop, increasing mRNA production of the SLC gene (as it was upregulated). Whilst the four animals with clinical signs of oxalate nephrosis show high expression, it is, however, not clear what effects the SNPs identified in this study have on the protein’s structure and function, or whether the primary defect is in this protein or another in the oxalate metabolic pathways, thereby driving an increased expression of SLC26A6 to increase oxalate secretion. Other genes of interest for the cause of oxalate nephrosis in koalas (Speight et al. 2018), including those that have been well described in primary hyperoxaluria in humans, are not expressed in lymph node tissue in other animals (Danpure and Rumsby 2004), and hence would likely not have been detected in the current study. If these genes were also involved in koalas with oxalate nephrosis, the SLC26A6 upregulation could possibly reflect a systemic response to increased oxalate load in SA koalas. Therefore, a targeted approach to investigating genes of interest and their expression is needed, with comparison between large cohorts of affected and non-affected koalas.
In the case of the marked propensity in the SA population to testicular abnormalities such as aplasia, the downregulated expression of the androgen receptor, a key gene in genital development and differentiation in mammals (Krausz 2011) and associated with male infertility in humans, gives an indication of which primary developmental pathways could be involved with this syndrome in koalas. It is possible that there is perturbation and compensatory increase in some genes in testicular development pathways due to aberrant decrease in critical regulators of sex differentiation such as the androgen receptor. Of critical note, there is no particular reason why these genes should be differentially expressed in lymph node tissue (which has little role in the functions of these genes), indicating that there may be a systemic (and not tissue specific) change in expression of these genes. None of the 12 male SA koalas in the study cohort were affected by testicular abnormality, hence further studies are needed in affected individuals to investigate expression of these genes in testis tissue.
The presence of scoliosis and kyphosis which has been reported in koalas in the SA population, and was present in two SA koalas in the study cohort, may be linked to the large number of genes associated with brain development as many genetic neuromuscular developmental disorders in humans present with scoliosis as part of the phenotype (Mary et al. 2018; Tully et al. 2018). However, the large number of genes differing between the populations in this study indicate that this phenotype is probably multifactorial in koalas. In addition the underlying defect (or defects) may not be in these genes per se but the regulatory networks controlling them, though in this respect it is important to note that transcription factors known to drive expression of these gene networks were not differentially expressed in this data set.
The “syncytin-1” like transcripts (Online resource 9), which are differentially expressed in different directions (2 upregulated and one down regulated in SA animals), are variants of a “type D” retroviral env gene (Henzy and Johnson 2013) and appear to be an endogenous retrovirus closely related to the previously described brush tailed possum (Trichosurus vulpeca) retrovirus TvERV (Online resource 9) (Baillie and Wilkins 2001). Up-regulation of human endogenous retroviral transcripts (HERVs) in response to exogenous retroviral infection has been well described for HIV infection of human cells (van der Kuyl 2012) and with the advent of long read deep sequencing capable of reliably distinguishing differing loci of HERV groups (there are up to several hundred of each type in a genome) it has become evident that HIV infection preferentially upregulates transcription from some loci while repressing others (Young et al. 2018), the mechanisms of which are not yet clear. It would not be unreasonable to expect that the high levels of KoRV expression in the QLD group (compared with the SA group) (Tarlinton et al. 2017) are affecting expression of other endogenous retroviruses in these animals similar to the effects HIV infection has on HERVs.
The results give an insight into the genetic basis of testicular abnormalities, oxalate nephrosis, and microcephaly syndromes seen in the genetically restricted SA populations and provide a starting point for targeted testing for the underlying genetic mutations causing these problems. They also provide a warning to managers of koala populations as to the types of developmental disorders that may be seen with further restriction of this population, such as might occur with groups used as founder populations in new areas.
There are limitations when working with a wild population which would ideally be addressed in a more targeted study. A definitive genetic study for SNPs associated with disease in these populations should involve a genome wide association study of DNA, a more carefully controlled “case” population and a much larger sample set than in this study. Case and control sample sizes in similar human studies of complex diseases are typically in the thousands (Spencer et al. 2009). However, there are a number of practical difficulties in achieving this in a vulnerable wildlife species where sampling is by necessity opportunistic. Given these limitations, the data provided here provides clear indication of which metabolic pathways may be involved in or altered by heritable genetic disorders in SA koalas and will provide a valuable starting point for population managers interested in the range of genetic diversity seen in different koala populations in Australia. In addition, the recent bushfire crisis with its subsequent effects of a drastic reduction in koala numbers has provided clear urgency to understanding which animals and populations are best suited to breeding and translocation programmes to maintain genetic diversity and population health in this vulnerable species.
Availability of data and material
Raw sequence reads fastq format are available at ENA with the accession number [PRJEB21505]. Data on animal demographics is provided in Online resource 1.
Code availability
Access details for all software used in this project is provided where appropriate in the methods (the software used in this paper is freely available or commercial software).
References
Andrews KR, Good JM, Miller MR, Luikart G, Hohenlohe PA (2016) Harnessing the power of RADseq for ecological and evolutionary genomics. Nat Rev Genet 17:81–92
Australian Senate Committees (2011) The koala, saving our national icon. In: Senate committee reports. Senate Printing Unit, Parliament House Canberra
Baillie GJ, Wilkins RJ (2001) Endogenous type D retrovirus in a marsupial, the common brushtail possum, (Trichosurus vulpecula). J Virol 75:2499–2507
BCFTOOLS. https://github.com/samtools/bcftools/
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Ser B 57:289–300
Bergin TJ (1978) In: The koala: proceedings of the taronga symposium on koala biology, management, and medicine, Sydney 11th and 12th March 197. 1978. Zoological Parks Board of NSW
Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527
Cammen KM, Andrews KR, Carroll EL, Foote AD, Humble E, Khudyakov JI, Louis M, McGowen MR, Olsen MT, van Cise AM (2016) Genomic methods take the plunge: recent advances in high-throughput sequencing of marine mammals. J Hered 107:481–495
Canfield PJ, Hartley WJ, Reddacliff GL (1990) Spontaneous proliferations in Australian marsupials—a survey and review. 1 macropods, koalas, wombats, possums and gliders. J Comp Pathol 103:135–145
Chen J, Bardes EE, Aronow BJ, Jegga AG (2009) ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37:W305–W311
Cingolani P, Platts A, Le Wang L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6:80–92
Cristescu R, Cahill V, Sherwin WB, Handasyde K, Carlyon K, Whisson D, Herbert CA, Carlsson F, Wilton AN, Cooper DW (2009) Inbreeding and testicular abnormalities in a bottlenecked population of koalas (Phascolarctos cinereus). Wildl Res 36:2989–3308
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, Depristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, Mcvean G, Durbin R, 1000 Genomes Project Analysis Group (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
Danpure CJ, Rumsby G (2004) Molecular aetiology of primary hyperoxaluria and its implications for clinical management. Expert Rev Mol Med 6:1–16
Fabijan J, Caraguel C, Jelocnik M, Polkinghorne A, Boardman WSJ, Nishimoto E, Johnsson G, Molsher R, Woolford L, Timms P, Simmons G, Hemmatzadeh F, Trott DJ, Speight N (2019a) Chlamydia pecorum prevalence in South Australian koala (Phascolarctos cinereus) populations: Identification and modelling of a population free from infection. Sci Rep 9:6261
Fabijan J, Miller D, Olagoke O, Woolford L, Boardman W, Timms P, Polkinghorne A, Simmons G, Hemmatzadeh F, Trott DJ, Speight KN (2019b) Prevalence and clinical significance of koala retrovirus in two South Australian koala (Phascolarctos cinereus) populations. J Med Microbiol 68:1072–1080
Fabijan J, Sarker N, Speight N, Owen H, Meers J, Simmons G, Seddon J, Emes RD, Tarlinton R, Hemmatzadeh F, Woolford L, Trott DJ (2020) Pathological findings in koala retrovirus-positive koalas (Phascolarctos cinereus) from Northern and Southern Australia. J Comp Pathol 176:50–66
Fowler EV, Hoeben P, Timms P (1998) Randomly amplified polymorphic DNA variation in populations of eastern Australian koalas, Phascolarctos cinereus. Biochem Genet 36:381–393
Freel RW, Hatch M, Green M, Soleimani M (2006) Ileal oxalate absorption and urinary oxalate excretion are enhanced in Slc26a6 null mice. Am J Physiol Gastrointest Liver Physiol 290:G719–G728
Gienapp P, Fior S, Guillaume F, Lasky JR, Sork VL, Csillery K (2017) Genomic quantitative genetics to study evolution in the wild. Trends Ecol Evol 32:897–908
Hanger J, Loader J (2014) Disease in wild koalas (Phascolarctos cinereus) with possible koala retrovirus involvement. In: Pye GW, Johnson RN, Greenwood AD (eds) The koala and its retroviruses: implications for sustainability and survival, 2014 San Diego. Technical Reports of the Australian Museum, pp 19–29
Heiene R, Rumsby G, Ziener M, Dahl SA, Tims C, Teige J, Ottesen N (2009) Chronic kidney disease with three cases of oxalate-like nephrosis in Ragdoll cats. J Feline Med Surg 11:474–480
Henzy JE, Johnson WE (2013) Pushing the endogenous envelope. Philos Trans R Soc Lond B Biol Sci 368:20120506
Higgins DP, Rose K, Spratt D (2018) Monotremes and Marsupials. In: Terio KA, Mcaloose D, Leger J (eds) Pathology of wildlife and zoo animals. Academic Press, New York
Hindrikson M, Remm J, Pilot M, Godinho R, Stronen AV, Baltrūnaité L, Czarnomska SD, Leonard JA, Randi E, Nowak C, Åkesson M, López-Bao JV, Álvares F, Llaneza L, Echegaray J, Vilà C, Ozolins J, Rungis D, Aspi J, Paule L, Skrbinšek T, Saarma U (2017) Wolf population genetics in Europe: a systematic review, meta-analysis and suggestions for conservation and management. Biol Rev Camb Philos Soc 92:1601–1629
Houlden BA, Greville WD, Sherwin WB (1996) Evolution of MHC class I loci in marsupials: characterization of sequences from koala (Phascolarctos cinereus). Mol Biol Evol 13:1119–1127
Houlden BA, Costello BH, Sharkey D, Fowler EV, Melzer A, Ellis W, Carrick F, Baverstock PR, Elphinstone MS (1999) Phylogeographic differentiation in the mitochondrial control region in the koala, Phascolarctos cinereus (Goldfuss 1817). Mol Ecol 8:999–1011
Jiang Z, Asplin JR, Evan AP, Rajendran VM, Velazquez H, Nottoli TP, Binder HJ, Aronson PS (2006) Calcium oxalate urolithiasis in mice lacking anion transporter Slc26a6. Nat Genet 38:474–478
Jiang H, Lei R, Ding SW, Zhu S (2014) Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15:182
Jiang H, Pokhrel G, Chen Y, Wang T, Yin C, Liu J, Wang S, Liu Z (2018) High expression of SLC26A6 in the kidney may contribute to renal calcification via an SLC26A6-dependent mechanism. PeerJ 6:e5192
Jones MR, Good JM (2016) Targeted capture in evolutionary and ecological genomics. Mol Ecol 25:185–202
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915
Kjeldsen SR, Raadsma HW, Leigh KA, Tobey JR, Phalen D, Krockenberger A, Ellis WA, Hynes E, Higgins DP, Zenger KR (2018) Genomic comparisons reveal biogeographic and anthropogenic impacts in the koala (Phascolarctos cinereus): a dietary-specialist species distributed across heterogeneous environments. Heredity 122:525–544
Köhler S, Carmody L, Vasilevsky N, Jacobsen JOB, Danis D, Gourdine JP, Gargano M, Harris NL, Matentzoglu N, McMurry JA, Osumi-Sutherland D, Cipriani V, Balhoff JP, Conlin T, Blau H, Baynam G, Palmer R, Gratian D, Dawkins H, Segal M, Jansen AC, Muaz A, Chang WH, Bergerson J, Laulederkind SJF, Yüksel Z, Beltran S, Freeman AF, Sergouniotis PI, Durkin D, Storm AL, Hanauer M, Brudno M, Bello SM, Sincan M, Rageth K, Wheeler MT, Oegema R, Lourghi H, Della Rocca MG, Thompson R, Castellanos F, Priest J, Cunningham-Rundles C, Hegde A, Lovering RC, Hajek C, Olry A, Notarangelo L, Similuk M, Zhang XA, Gómez-Andrés D, Lochmüller H, Dollfus H, Rosenzweig S, Marwaha S, Rath A, Sullivan K, Smith C, Milner JD, Leroux D, Boerkoel CF, Klion A, Carter MC, Groza T, Smedley D, Haendel MA, Mungall C, Robinson PN (2019) Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res 47:D1018–D1027
Krausz C (2011) Male infertility: pathogenesis and clinical diagnosis. Best Pract Res Clin Endocrinol Metab 25:271–285
Lau Q, Jaratlerdsiri W, Griffith JE, Gongora J, Higgins DP (2014) MHC class II diversity of koala (Phascolarctos cinereus) populations across their range. Heredity 113:287–296
Legione AR, Patterson JL, Whiteley P, Firestone SM, Curnick M, Bodley K, Lynch M, Gilkerson JR, Sansom FM, Devlin JM (2016) Koala retrovirus (KoRV) genotyping analyses reveal a low prevalence of KoRV-A in Victorian koalas and an association with clinical disease. J Med Microbiol 66:236–244
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009) The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25:2078–2079
Lindsay HA (1950) Re-establishing the koala in SA. Wild Life 12:257–262
Lu X, Sun D, Xu B, Pan J, Wei Y, Mao X, Yu D, Liu H, Gao B (2016) In Silico screening and molecular dynamic study of nonsynonymous single nucleotide polymorphisms associated with kidney stones in the SLC26A6 gene. J Urol 196:118–123
Martin R, Handasyde K (1999) The koala: natural history, conservation and management. University of New South Wales Press, Sydney
Mary P, Servais L, Vialle R (2018) Neuromuscular diseases: diagnosis and management. Orthop Traumatol Surg Res 104:S89-s95
Monico CG, Weinstein A, Jiang Z, Rohlinger AL, Cogal AG, Bjornson BB, Olson JB, Bergstralh EJ, Milliner DS, Aronson PS (2008) Phenotypic and functional analysis of human SLC26A6 variants in patients with familial hyperoxaluria and calcium oxalate nephrolithiasis. Am J Kidney Dis 52:1096–1103
Neaves LE, Frankham GJ, Dennison S, Fitzgibbon S, Flannagan C, Gillett A, Hynes E, Handasyde K, Helgen KM, Tsangaras K, Greenwood AD, Eldridge MD, Johnson RN (2016) Phylogeography of the koala, (Phascolarctos cinereus), and harmonising data to inform conservation. PLoS ONE 11:e0162207
O’Brien SJ, Johnson WE, Driscoll CA, Dobrynin P, Marker L (2017) Conservation genetics of the cheetah: lessons learned and new opportunities. J Hered 108:671–677
Oikkonen L, Lise S (2017) Making the most of RNA-seq: pre-processing sequencing data with Opossum for reliable SNP variant detection. Wellcome Open Res 17:6
Pimentel HJ, Bray N, Puente S, Melsted P, Pachter L (2017) Differential analysis of RNA-Seq incorporating quantification uncertainty. Nat Methods 14:687–690
Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, Furlong LI (2020) The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res 48(D1):D845–D855
Polkinghorne A, Hanger J, Timms P (2013) Recent advances in understanding the biology, epidemiology and control of chlamydial infections in koalas. Vet Microbiol 165:214–223
Ruiz-Rodriguez CT, Ishida Y, Murray ND, O’Brien SJ, Graves JA, Greenwood AD, Roca AL (2016) Koalas (Phascolarctos cinereus) from Queensland are genetically distinct from 2 populations in Victoria. J Hered 107:573–580
Saremi NF, Supple MA, Byrne A, Cahill JA, Coutinho LL, Dalen L, Figueiro HV, Johnson WE, Milne HJ, O’Brien SJ, O’Connell B, Onorato DP, Riley SPD, Sikich JA, Stahler DR, Villela PMS, Vollmers C, Wayne RK, Eizirik E, Corbett-Detig RB, Green RE, Wilmers CC, Shapiro B (2019) Puma genomes from North and South America provide insights into the genomic consequences of inbreeding. Nat Commun 10:4769
Sarker N, Fabijan J, Owen H, Seddon J, Simmons G, Speight K, Kaler J, Woolford L, Emes RD, Hemmatzadeh F, Trott D, Meers J, Tarlinton R (2019) Koala retrovirus viral load and disease burden in distinct northern and southern koala populations. Sci Rep (In press)
Seymour AM, Montgomery ME, Costello BH, Ihle S, Johnsson G, St John B, Taggart D, Houlden BA (2001) High effective inbreeding coefficients correlate with morphological abnormalities in populations of South Australian koalas (Phascolarctos cinereus). Anim Conserv Forum 4:211–219
Simmons GS, Young PR, Hanger JJ, Jones K, Clarke D, McKee JJ, Meers J (2012) Prevalence of koala retrovirus in geographically diverse populations in Australia. Aust Vet J 90:404–409
Speight KN, Boardman W, Breed WG, Taggart DA, Woolford L, Haynes JI (2013) Pathological features of oxalate nephrosis in a population of koalas (Phascolarctos cinereus) in South Australia. Vet Pathol 50:299–307
Speight KN, Haynes JI, Boardman W, Breed WG, Taggart DA, Rich B, Woolford L (2014) Plasma biochemistry and urinalysis variables of koalas (Phascolarctos cinereus) with and without oxalate nephrosis. Vet Clin Pathol 43:244–254
Speight KN, Hicks P, Graham C, Boardman W, Breed WG, Manthorpe E, Funnell O, Woolford L (2018) Necropsy findings of koalas from the Mount Lofty Ranges population in South Australia. Aust Vet J 96:188–192
Spencer CCA, Su Z, Donnelly P, Marchini J (2009) Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet 5:e1000477–e1000477
Steiner CC, Putnam AS, Hoeck PE, Ryder OA (2013) Conservation genomics of threatened animal species. Annu Rev Anim Biosci 1:261–281
Sutton RH (1986) Craniofacial tumors of the koala (Phascolarctos cinereus). J Wildl Dis 22:283–285
Tarlinton R, Sarker N, Fabijan J, Dottorini T, Woolford L, Meers J, Simmons G, Owen H, Seddon J, Hemmatzedah F, Trott D, Speight N, Emes RD (2017) Differential and defective expression of Koala Retrovirus reveal complexity of host and virus evolution. BioRxiv. https://doi.org/10.1101/211466v1.abstract
Taylor AC, Graves JM, Murray ND, O’Brien SJ, Yuhki N, Sherwin B (1997) Conservation genetics of the koala (Phascolarctos cinereus): low mitochondrial DNA variation amongst southern Australian populations. Genet Res 69:25–33
Tully PA, Edwards BA, Mograby O, Davis HSM, Arieskola O, Magdum S, Rao P, Jayamohan J (2018) Should all paediatric patients with presumed idiopathic scoliosis undergo MRI screening for neuro-axial disease? Childs Nerv Syst 34:2173–2178
Van Der Kuyl AC (2012) HIV infection and HERV expression: a review. Retrovirology 9:6
Vidgren G, Vainio-Siukola K, Honkasalo S, Dillard K, Anttila M, Vauhkonen H (2012) Primary hyperoxaluria in Coton de Tulear. Anim Genet 43:356–361
Wier B, Cockerham C (1984) Estimating statistics for the analysis of population structure. Evolution 38:1358–1370
Woinarski J, Burbidge AA (2016) Phascolarctos cinereus. The IUCN Red List of Threatened Species 2016, e.T16892A21960344
Young GR, Terry SN, Manganaro L, Cuesta-Dominguez A, Deikus G, Bernal-Rubio D, Campisi L, Fernandez-Sesma A, Sebra R, Simon V, Mulder LCF (2018) HIV-1 infection of primary CD4(+) T cells regulates the expression of specific human endogenous retrovirus HERV-K (HML-2) elements. J Virol. https://doi.org/10.1128/JVI.01507-17
Acknowledgements
The authors thank Dr Mark Ravinet, School of Life Sciences, University of Nottingham for advice on statistical analysis”
Funding
This project was funded by the Queensland Department of the Environment and Heritage Koala Research Grant Programme 2012. NSa was also supported by a Keith Mackie Lucas travel scholarship from the University of Queensland.
Author information
Authors and Affiliations
Contributions
RET, JM, GS, JMS, HO, NSp, FH, RDE conceived and designed the study, RT wrote the paper, all authors provided critical commentary, RDE generated the figures, NSa, JF, LW, HO, NSp and RDE performed sample collection, sample processing and or data analysis.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no competing interests or conflicts of interest.
Consent to participate
Koalas for post mortem were accessed through the Moggill Wildlife hospital (QLD Department of the Environment and Heritage Protection) and the Adelaide Koala and Wildlife Hospital, Plympton, South Australia and Fauna Rescue of South Australia Inc.
Consent for publication
All authors consent to publication.
Ethical approval
Ethical approval for this study was granted by the University of Queensland Animal Ethics Committee, permit number ANFRA/SVS/461/12 and ANRFA/SVS/445/15, the Queensland Government Department of Environment and Heritage Protection permit number WISP11989112, the University of Adelaide Animal Ethics Committee permit number S-2013-198 and the South Australian Government Department of Environment, Water and Natural Resources Scientific Research Permit Y26054.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10592_2021_1340_MOESM1_ESM.xlsx
Electronic supplementary material 1 (XLSX 16 kb) Online resource 1: Details of the animals included in this study: N/A = sample not available for testing, region of origin, QLD= Queensland, SA= South Australia, sex, M= Male, F= female, tooth wear class (age classification on a 7 point scale), pathology, KoRV proviral status (pol gene PCR on DNA from whole blood), lymph node RNA quality and NGS read details: concentration, A260/280 and A260/230 ratios, RIN value, number of paired raw reads and number of trimmed reads from each sample.
10592_2021_1340_MOESM2_ESM.xlsx
Electronic supplementary material 2 (XLSX 12915 kb) Online resource 2: SNPs with FST greater or equal than 0.8, functional annotations. Gene ID, chromosomal location, sequence in reference genome, alternate SNP in this dataset, presence or absence in individual animals (./. indicates transcript location not covered in that animal, 0/1= SNP heterozygous, 1/1 = SNP homozygous in that animal) , proportion of transcript not covered, type of change (SNP or indel or alternate transcript, stop codon or UTR variant), effect of change (synonymous/non-synonymous, Impact on gene function (modifier, low, high, moderate), position of change, amino acid change, Gene ID (description and identifier)
10592_2021_1340_MOESM3_ESM.xlsx
Electronic supplementary material 3 (XLSX 1375 kb) Online resource 3: Genes associated with brain abnormalities functional annotations Gene ID, chromosomal location, sequence in reference genome, alternate SNP in this dataset, presence or absence in individual animals (./. indicates transcript location not covered in that animal, 0/1= SNP heterozygous, 1/1 = SNP homozygous in that animal) , proportion of transcript not covered, type of change (SNP or indel or alternate transcript, stop codon or UTR variant), effect of change (synonymous/non-synonymous, Impact on gene function (low, high, moderate), position of change, amino acid change, Gene ID (description and identifier)
10592_2021_1340_MOESM4_ESM.xlsx
Electronic supplementary material 4 (XLSX 25 kb) Online resource 4: Genes in pathways associated with brain disease, Pathway ID, name, P values, q values (Bonferrroni and FDR corrected) , number of genes in pathway from this data set (and total number in genome), Gene IDs of genes from this data set in pathway
10592_2021_1340_MOESM5_ESM.xlsx
Electronic supplementary material 5 (XLSX 82 kb) Online resource 5: Differentially expressed genes between SA and QLD animals
10592_2021_1340_MOESM6_ESM.xlsx
Electronic supplementary material 6 (XLSX 55 kb) Online resource 6: SNPs with FST greater or equal than 0.8 associated with differentially expressed genes between SA and QLD animals
10592_2021_1340_MOESM7_ESM.pdf
Electronic supplementary material 7 (PDF 301 kb) Online resource 7: Amino acid sequence data of QLD and SA variants of SLC26A6 transcript variant XM_020999000.1.
10592_2021_1340_MOESM8_ESM.txt
Electronic supplementary material 8 (TXT 18 kb) Online resource 8: Fasta file of sequences of transcript variants of SLC26A6
10592_2021_1340_MOESM9_ESM.pdf
Electronic supplementary material 9 (PDF 294 kb) Online resource 9: Analysis of the Type D env retroviral sequences detected in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tarlinton, R.E., Fabijan, J., Hemmatzadeh, F. et al. Transcriptomic and genomic variants between koala populations reveals underlying genetic components to disorders in a bottlenecked population. Conserv Genet 22, 329–340 (2021). https://doi.org/10.1007/s10592-021-01340-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-021-01340-7