Abstract
A novel bacterium capable of utilizing 2-sec-butylphenol as the sole carbon and energy source, Pseudomonas sp. strain MS-1, was isolated from freshwater sediment. Within 30 h, strain MS-1 completely degraded 1.5 mM 2-sec-butylphenol in basal salt medium, with concomitant cell growth. A pathway for the metabolism of 2-sec-butylphenol by strain MS-1 was proposed on the basis of the identification of 3 internal metabolites—3-sec-butylcatechol, 2-hydroxy-6-oxo-7-methylnona-2,4-dienoic acid, and 2-methylbutyric acid—by gas chromatography-mass spectrometry analysis. Strain MS-1 degraded 2-sec-butylphenol through 3-sec-butylcatechol along a meta-cleavage pathway. Degradation experiments with various alkylphenols showed that the degradability of alkylphenols by strain MS-1 depended strongly on the position (ortho ≫ meta = para) of the alkyl substitute, and that strain MS-1 could degrade 2-alkylphenols with various sized and branched alkyl chain (o-cresol, 2-ethylphenol, 2-n-propylphenol, 2-isopropylphenol, 2-sec-butylphenol, and 2-tert-butylphenol), as well as a dialkylphenol (namely, 6-tert-butyl-m-cresol).
Similar content being viewed by others
Introduction
Alkylphenols, a group of chemical compounds that consist of a phenol with an alkyl chain, are ubiquitous pollutants in urban aquatic environments. Alkylphenols are industrially important chemicals and are abundantly and widely used to produce alkylphenol polyethoxylates and phenolic resins (McLeese et al. 1981; Ying et al. 2002). Furthermore, alkylphenols with an alkyl chain longer than 3 carbon atoms (≥C3) interact with estrogen receptors (Hu and Aizawa 2003; Routledge and Sumpter 1997; Tollefsen et al. 2008; White et al. 1994) and have other toxic effects on aquatic organisms and humans (Mcleese et al. 1981; Servos 1999). Therefore, considerable attention has been paid to their environmental fates and biodegradabilities.
Studies of the biodegradation of alkylphenols have focused mainly on para-substituted alkylphenols, including 4-nonylphenol, 4-octylphenol, and 4-alkylphenol with medium-length alkyl chains (C3 to C7). Several sphingomonads, including Sphingomonas sp. strain TTNP3 (Tanghe et al. 1999), Sphingobium xenophagum Bayram (Gabriel et al. 2005), and Sphingomonas cloacae (Fujii et al. 2001), have recently been isolated from activated sludge; these sphingomonads can degrade branched 4-nonylphenol and utilize it as a sole carbon source. In addition, several Pseudomonas strains that can degrade medium-length 4-n-alkylphenol and utilize it as a sole carbon source, including Pseudomonas sp. KL28 (Jeong et al. 2003), Pseudomonas putida MT4 (Takeo et al. 2006), and Pseudomonas veronii INA06 (Ajithkumar et al. 2003), have been isolated from activated sludge or contaminated soil. In contrast, there has been only one previously reported bacterial strain capable of degrading ortho-substituted alkylphenols (2-alkylphenols) with an alkyl chain of ≥C3, including 2-n-propylphenol, 2-isopropylphenol, and 2-sec-butylphenol; namely, Pseudomonas azelaica HBP1 Prp (Kohler et al. 1993; Reichlin and Kohler 1994; van der Maarel and Kohler 1993), which is a regulatory mutant that grows on 2-propylphenol and 2-sec-butylphenol. The wild-type strain HBP1 cannot grow on 2-alkylphenol (Kohler et al. 1993).
Therefore, it remains unclear which bacteria can degrade 2-alkylphenols and how this biodegradation occurs in the natural aquatic environment. Because 2-alkylphenols with an alkyl chain of ≥C3, including 2-propylphenols and 2-butylphenols, exhibit estrogenic activity (Hu and Aizawa 2003; Routledge and Sumpter 1997; Tollefsen et al. 2008) and other toxic effects (Acuña-Argüelles et al. 2003; Choi et al. 2004; Li et al. 2008), it is important to understand their fates and biodegradation in the aquatic environment.
Here we report the characterization of Pseudomonas sp. strain MS-1 isolated from a natural freshwater sediment. The strain was capable of utilizing 2-sec-butylphenol as the sole carbon source.
Materials and methods
Chemicals
Phenol, o-cresol, m-cresol, p-cresol, 2-ethylphenol, 3-ethylphenol, 4-ethylphenol, 2-n-propylphenol, 2-isopropylphenol, 3-isopropylphenol, 4-n-propylphenol, 4-isopropylphenol, 2-sec-butylphenol, 2-tert-butylphenol, 3-tert-butylphenol, 4-n-butylphenol, 4-sec-butylphenol, 4-tert-butylphenol, 2,4-di-tert-butylphenol, 2,6-di-tert-butylphenol, 6-tert-butyl-m-cresol, and N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) were purchased from Tokyo Chemical Industry (Tokyo, Japan). 2-Methylbutyric acid was purchased from Acros Organics (Morris Plains, New Jersey, USA).
Culture media
Basal salts medium (BSM) (Toyama et al. 2009) was used for degradation tests, and BSM containing 2-sec-butylphenol (2sBP-BSM) as the sole carbon source was used for culture of 2-sec-butylphenol-degrading bacteria. Agar solid medium was prepared with 2.0% (w/v) agar.
Enrichment, isolation, and identification of 2-sec-butylphenol—degrading bacteria
A sediment sample was obtained from within Phragmites australis (reed) zone in Inukai Pond at Osaka University, Suita, Osaka, Japan. The sediment sample was collected from about 20 cm sediment depth, around the plant roots. Detailed physicochemical analysis revealed that the sediment sample had a pH of 6.9, a low organic carbon content (ignition loss = 2.2%), and no alkylphenols (2-sec-butylphenol, 4-n-butylphenol, 4-tert-butylphenol, 4-tert-octylphenol, or nonylphenol). Enrichment of 2-sec-butylphenol-degrading bacteria from the sediment was performed by transferring 1 g of sediment sample to a 300-ml flask containing 100 ml 2sBP-BSM (0.5 mM). The mixture was incubated at 28°C on a rotary shaker at 120 rpm for 14 days. We then transferred 1 ml of the culture of sediment to fresh 2sBP-BSM (0.5 mM). After third transfers, the enrichment culture was serially diluted and spread on 2sBP-BSM (0.5 mM) agar plates, and the plants were incubated at 28°C. The isolated bacterial strain, designated MS-1, was characterized and identified by physiological and phylogenetic analyses (Inoue et al. 2008).
Growth and degradation experiments
Strain MS-1 was grown overnight in 2sBP-BSM (1.0 mM). Cells were harvested by centrifugation (9,600g, 4°C, 10 min), washed twice with 50 mM potassium phosphate buffer (pH 7.5), and inoculated at a cell density of 0.02 (i.e., optical density at a wavelength of 600 nm [OD600] = 0.02) into 2sBP-BSM (1.5 mM). Also, the cells were inoculated at an OD600 of 0.02 into BSM containing 1.5 mM 2-methylbutyric acid as the sole carbon source. Cultivation was carried out at 28°C and 120 rpm. Cell densities, concentrations of 2-sec-butylphenol and 2-methylbutyric acid, and metabolites produced by 2-sec-butylphenol degradation were monitored periodically over the 48-h experimental period.
Alkylphenols with alkyl chains of various sizes, branches, and positions were used for growth experiments on strain MS-1 and for degradation tests using whole-cell strain MS-1. Cells, prepared as described above, were added at an OD600 of 0.02 (in the growth experiments) or 0.5 (in the degradation tests) to BSM supplemented with the following alkylphenols as the sole carbon source at 0.5 mM: phenol, o-cresol, m-cresol, p-cresol, 2-ethylphenol, 3-ethylphenol, 4-ethylphenol, 2-n-propylphenol, 2-isopropylphenol, 3-isopropylphenol, 4-n-propylphenol, 4-isopropylphenol, 2-sec-butylphenol, 2-tert-butylphenol, 3-tert-butylphenol, 4-n-butylphenol, 4-sec-butylphenol, 4-tert-butylphenol, 2,4-di-tert-butylphenol, 2,6-di-tert-butylphenol, and 6-tert-butyl-m-cresol. Cultivation was performed as 28°C and 120 rpm. Cell densities and substrate concentrations were monitored every 12 h over the 48-h experimental period.
Analytical procedures
Bacterial growth was monitored by recording OD600, and the dry weight of the cells was determined. The cells were harvested by centrifugation (9,600g, 4°C, 10 min) and washed twice with 50 mM potassium phosphate buffer (pH 7.5), and the cell suspension was filtered through a pre-weighed filter (pore size 0.2 μm, polycarbonate; Millipore, Tokyo, Japan). The filter, together with the cells, was dried at 90°C for 3 h before weighing. Alkylphenol concentrations were determined by high-performance liquid chromatography (HPLC); the 2-methylbutyric acid concentration was determined by gas chromatography-mass spectrometry (GC-MS); and metabolites of 2-sec-butylphenol were analyzed by GC-MS. For the HPLC and GC-MS analyses, the collected culture was acidified with 1N HCl, shaken for 3 min with an equal volume of 1:1 (v:v) ethyl acetate:n-hexane, and centrifuged (9,600g, 4°C, 10 min); the organic layer was then extracted. The extract was directly analyzed by HPLC and GC-MS. Also, for GC-MS analysis of metabolites produced by 2-sec-butylphenol degradation, the extract was dried under a nitrogen flow and subjected to trimethylsilylation (TMS) using a BSTFA–acetonitrile solution at 60°C for 1 h. The HPLC analysis was conducted with a Shimadzu HPLC system with an SPD-10AV UV–vis detector and a Shim-pack VP-ODS column (4.5 × 150 mm, particle size 5 μm; Shimadzu, Kyoto, Japan). In the HPLC analysis, 8:2 (v:v) acetonitrile:water was used as a mobile phase, and the detection wavelength was 277 nm. The GC-MS analysis was conducted with a Shimadzu GC/MS system (GCMS-QP2010) and an Rxi-5 ms capillary column (30 m, 0.25 mm ID, 1.00 μm df; Restek, Pennsylvania, USA). In the GC-MS analysis, the column temperature programs were as follows: (i) hold at 90°C for 1 min, increase to 150 at 15°C min−1, increase to 300°C at 5°C min−1, and hold at 300°C for 5 min for analysis of metabolites with TMS derivatization; and (ii) hold at 60°C for 2 min, increase to 300°C at 20°C min−1, and hold at 300°C for 5 min for analysis of metabolites without TMS derivatization. The injection, interface, and ion-source temperatures were 280, 280, and 250°C, respectively. Helium (99.995%) was used as the carrier gas at a flow rate of 1.0 ml min−1.
Nucleotide sequence accession number
The 16S rRNA gene sequence of strain MS-1 has been submitted to the DDBJ/EMBL/GenBank databases under accession no. AB499847.
Results
Isolation and identification of 2-sec-butylphenol-degrading bacteria
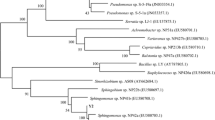
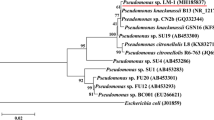
The sediment sample was incubated in 2sBP-BSM (0.5 mM) to check its 2-sec-butylphenol-degrading ability. The 2-sec-butylphenol completely disappeared from the sediment culture within 14 days. Enrichment culture of the sediment sample resulted in the isolation of a bacterial strain, MS-1, that could be aerobically grown on 0.5 mM 2-sec-butylphenol as the sole carbon and energy source. Strain MS-1 formed cream colonies on 2sBP-BSM (0.5 mM) agar. The isolate was a rod-shaped, gram-negative, catalase- and oxidase-positive bacterium. The isolate utilized glucose, d-mannose, gluconate, n-caprate, d,l-malate, citrate, and phenylacetate as sole carbon sources, but not l-arabinose, d-mannitol, N-acetyl-d-glucosamine, maltose, or adipate. The partial 16S rRNA gene sequence of the isolate showed the closest sequence identity (99.0%) to Pseudomonas taiwanensis BCRC 17751T (accession no. EU103629). Therefore, we identified strain MS-1 as a strain of Pseudomonas sp. Figure 1 shows the phylogenetic relationships that were established on the basis of the 16S rRNA gene sequence of strain MS-1. Figure 1 also shows closely related type strains and previously isolated alkylphenol-degrading strains. Our isolate was phylogenetically related to medium-length 4-n-alkylphenol-degrading bacteria, including Pseudomonas sp. CF600 (accession no. DQ777732), Pseudomonas putida MT4 (accession no. AB180734), and Pseudomonas sp. KL28 (accession no. AY324319).
Phylogenetic relationships established by the neighbor-joining method, based on 16S rRNA gene sequences of strain MS-1, type strains of Pseudomonas sp., and previously isolated medium-length 4-n-alkylphenol-degrading (*) bacterial strains. Numbers on branches indicate bootstrap confidence estimates obtained with 1,000 replicates. Scale bar represents an evolutionary distance (Knuc) of 0.01
Degradation of 2-sec-butylphenol by strain MS-1
Strain MS-1 pre-grown on 2-sec-butylphenol completely degraded 1.5 mM 2-sec-butylphenol within 30 h, and the bacterial cell density (OD600) increased in parallel with 2-sec-butylphenol degradation (Fig. 2). Yield coefficient of the cells was 68.5 mg dry weight/mmol of 2-sec-butylphenol (0.457 mg dry weight/mg of 2-sec-butylphenol). 2-Methylbutyric acid was detected by GC-MS analysis after 3 h of cultivation, along with decrease in 2-sec-butylphyenol. Retention time and MS spectrum of 2-methylbutyric acid corresponded to those of its authentic compound. The concentration of 2-methybutyric acid increased from 3 to 21 h of cultivation and reached a maximum of 0.13 mM after 21 h of cultivation, subsequently decreasing to an undetected level within 30 h (Fig. 2). In addition, strain MS-1 pre-grown on 2-sec-butylphenol rapidly and completely degraded 1.5 mM 2-methylbutyric acid within 12 h, with concomitant cell growth (data not shown). Yield coefficient of the cells was 39.5 mg dry weight/mmol of 2-methylbutyric acid (0.387 mg dry weight/mg of 2-methylbutyric acid).
Degradation of 2-sec-butylphenol and cell growth of strain MS-1 grown in basal salt medium (BSM) containing 1.5 mM 2-sec-butylphenol. Concentrations of 2-sec-butylphenol (closed squares) and 2-methylbutyric acid (open circles), and cell densities (open diamonds) were monitored over 48 h. Data represent the means of triplicate experiments, and error bars indicate 95% confidence intervals
Identification of metabolites and 2-sec-butylphenol degradation pathway
Analysis of the acetate:n-hexane extract with and without TMS derivatization by GC-MS showed the presence of 3-sec-butylcatechol (Fig. 3a) and its meta-cleavage product, namely, 2-hydroxy-6-oxo-7-methylnona-2,4-dienoic acid (Fig. 3b). MS spectra of 3-sec-butylcatechol and 2-hydroxy-6-oxo-7-methylnona-2,4-dienoic acid corresponded to those in previous report (van der Maarel and Kohler 1993). 3-sec-Butylcatechol and 2-hydroxy-6-oxo-7-methylnona-2,4-dienoic acid appeared and increased from 3 to 21 h of cultivation, and subsequently both metabolites completely disappeared within 30 h.
Mass spectra of metabolites formed during degradation of 2-sec-butylphenol by strain MS-1. a Metabolite I was identified as 3-sec-butylcatechol. b Metabolite II was identified as a meta-cleavage product, namely, 2-hydroxy-6-oxo-7-methylnona-2,4-dienoic acid, with trimethylsilylation (TMS) derivatization
On the basis of the metabolites identified and the results of previous study (van der Maarel and Kohler 1993), we propose a pathway for the degradation of 2-sec-butylphenol by strain MS-1 (Fig. 4). Briefly, 2-sec-butylphenol is hydroxylated to 3-sec-butylcatechol. Subsequently, 3-sec-butylcatechol is metabolized to 2-hydroxy-6-oxo-7-methylnona-2,4-dienoic acid. The meta-cleavage product further metabolized to 2-methylbutyric acid and, presumably, 2-hydroxypent-2,4-dienoic acid. The 2-methylbutyric acid is mineralized.
Utilization and degradability of various alkylphenols
Utilization and degradation of alkylphenols by strain MS-1 are summarized in Table 1. Among the alkylphenols tested, 2-n-propylphenol, 2-isopropylphenol, 2-tert-butylphenol, and 6-tert-butyl-m-cresol were utilized by strain MS-1 for growth, in addition to 2-sec-butylphenol. Whole cells pre-grown on 2-sec-butylphenol degraded substantial amounts of the following alkylphenols (final biotransformation ratio [%] within 48 h): o-cresol (100%) and 2-ethylphenol (100%). In contrast, phenol, m-cresol, p-cresol, 3-ethylphenol, 4-ethylphenol, 3-n-propylphenol, 3-isopropylphenol, 4-n-propylphenol, 4-isopropylphenol, 3-tert-butylphenol, 4-n-butylphenol, 4-sec-butylphenol, 4-tert-butylphenol, 2,4-di-tert-butylphenol, and 2,6-di-tert-butylphenol were neither utilized for growth nor degraded by MS-1. These results suggest that alkylphenol degradation by MS-1 depends on the position (ortho ≫ meta = para) of the alkyl chain and not on the branching and size of the alkyl chain.
Discussion
This study reports the isolation and characterization of an aerobic bacterium, Pseudomonas sp. strain MS-1, capable of utilizing 2-sec-butylphenol as the sole carbon and energy source. To our knowledge, strain MS-1 is the first 2-sec-butylphenol-utilizing bacterium isolated from a natural aquatic environment. Our strain MS-1 was phylogenetically related to medium-length 4-n-alkylphenol-degrading Pseudomonas strains, including strains CF600, MT4, KL28, and INA06, rather than to Pseudomonas azelaica strain HBP1 (Fig. 1).
Strain MS-1 degraded 1.5 mM 2-sec-butylphenol within 30 h, whereas strain HBP1 Prp can degrade 2.0 mM 2-sec-butylphenol within 45 h (van der Maarel and Kohler 1993), suggesting that the rate of 2-sec-butylphenol degradation by our strain MS-1 is comparable to that by strain HBP1 Prp. Our results indicated that the metabolism of 2-sec-butylphenol by strain MS-1 proceeds by the same metabolic pathway as degradation of 2-sec-butylphenol by strain HBP1 Prp (van der Maarel and Kohler 1993). That is, 2-sec-butylphenol is initially hydroxylated to 3-sec-butylcatechol, and 3-sec-butylcatechol is further metabolized along a meta-cleavage pathway (Fig. 4).
Our experiments show that the alkylphenol-metabolizing system in strain MS-1 has substrate specificity that depended on the position of the alkyl chain. Strain MS-1 could degrade ortho-butylphenols but not meta- and para-butylphenols. In contrast, strain MS-1 showed degradative activity for 2-alkylphenols with various sized and branched alkyl chain (methyl, ethyl, n-propyl, isopropyl, sec-butyl, and tert-butyl); the strain also utilized 2-alkylphenols with an alkyl chain (n-propyl, isopropyl, sec-butyl, and tert-butyl). Strain MS-1 also degraded a dialkylphenolic compound, namely, 6-tert-butyl-m-cresol and utilized it as the sole carbon source, but not 2,4-di-tert-butylphenol and 2,6-di-tert-butylphenol. Strain HBP1 Prp shows degradative activity for a variety of ortho-substituted alkylphenols with a short- or medium-length alkyl chain (o-cresol, 2-ethylphenol, 2-n-propylphenol, 2-isopropylphenol, and 2-sec-butylphenol), but it shows very little activity for 2-tert-butylphenol (Kohler et al. 1988, 1993). Thus, degradative range for 2-alkylphenols of strain MS-1 appears to be wider than that of strain HBP1 Prp.
Until now, nothing has been known about the bacteria that degrade ortho-substituted alkylphenols and the mechanisms of this biodegradation in the natural aquatic environment. Previously reported 4-NP-degrading (Tanghe et al. 1999) and medium-length 4-n-alkylphenol-degrading aerobic bacteria (Jeong et al. 2003; Takeo et al. 2006) cannot degrade ortho-substituted isomers. In contrast, strain MS-1 degraded a variety of alkylphenols only with orth-substituted alkyl chain. This interesting characteristic is very different from previously reported alkylphenol degraders. Our results provide new insight into the degradation of 2-alkylphenols and will contribute to remediation of alkylphenol pollutants generally in aquatic environments. Further study to gain a more detailed understanding of the 2-sec-butylphenol enzymes of strain MS-1 is under way.
References
Acuña-Argüelles ME, Olguin-Lora P, Razo-Flores E (2003) Toxicity and kinetic parameters of the aerobic biodegradation of the phenol and alkylphenols by a mixed culture. Biotechnol Lett 25:559–564. doi:10.1023/A:1022898321664
Ajithkumar B, Ajithkumar VP, Iriye R (2003) Degradation of 4-amylphenol and 4-hexylphenol by a new activated sludge isolate of Pseudomonas veronii and proposal for a new subspecies status. Res Microbiol 154:17–23. doi:10.1016/S0923-2508(02)00009-8
Choi K, Sweet LI, Meier PG, Kim P-G (2004) Aquatic toxicity of four alkylphenols (3-tert-butylphenol, 2-isopropylphenol, 3-isopropylphenol, and 4-isopropylphenol) and their binary mixtures to microbes, invertebrates, and fish. Environ Toxicol 19:45–50
Fujii K, Urano N, Ushio H, Satomi M, Kimura S (2001) Sphingomonas cloacae sp. nov., a nonylphenol-degrading bacterium isolated from wastewater of a sewage-treatment plant in Tokyo. Int J Syst Evol Microbiol 51:603–610
Gabriel FLP, Giger W, Guenther K, Kohler H-PE (2005) Differential degradation of nonylphenol isomers by Sphingomonas xenophaga Bayram. Appl Environ Microbiol 71:1123–1129. doi:10.1128/AEM.71.3.1123-1129.2005
Hu J-Y, Aizawa T (2003) Quantitative structure–activity relationships for estrogen receptor binding affinity of phenolic chemicals. Water Res 37:1213–1222
Inoue D, Hara S, Kashihara M, Murai Y, Danzl E, Sei K, Tsunoi S, Fujita M, Ike M (2008) Degradation of bis(4-hydroxyphenyl) methane (bisphenol F) by Sphingobium yanoikuyae strain FM-2 isolated from river water. Appl Environ Microbiol 74:352–358. doi:10.1128/AEM.01708-07
Jeong JJ, Kim JH, Kim C-K, Hwang I, Lee K (2003) 3- and 4-Alkylphenol degradation pathway in Pseudomonas sp. strain KL28: genetic organization of lap gene cluster and substrate specificities of phenol hydroxylase and catechol 2,3-dioxygenase. Microbiology 149:3265–3277. doi:10.1099/mic.0.26628-0
Kohler H-PE, Kohler-Staub D, Focht DD (1988) Degradation of 2-hydroxybiphenyl and 2,2′-dihydroxybiphenyl by Pseudomonas sp. strain HBP1. Appl Environ Microbiol 54:2683–2688
Kohler H-PE, van der Maarel MJEC, Kohler-Staub D (1993) Selection of Pseudomonas sp. strain HBP1 Prp for metabolism of 2-propylphenol and elucidation of the degradative pathway. Appl Environ Microbiol 59:860–866
Li J, Ma M, Wang Z (2008) A two-hybrid yeast assay to quantify the effects of xenobiotics on thyroid hormone-mediated gene expression. Environ Toxicol Chem 27:159–167. doi:10.1897/07-054.1
McLeese DW, Zitko V, Sergeant DB, Burridge L, Metcalfe CD (1981) Lethality and accumulation of alkylphenols in aquatic fauna. Chemosphere 10:723–730
Reichlin F, Kohler H-PE (1994) Pseudomonas sp. strain HBP1 Prp degrades 2-isopropylphenol (ortho-cumenol) via meta cleavage. Appl Environ Microbiol 60:4587–4591
Routledge EJ, Sumpter JP (1997) Structural features of alkylphenolic chemicals associated with estrogenic activity. J Biol Chem 272:3280–3288
Servos MR (1999) Review of the aquatic toxicity, estrogenic responses and bioaccumulation of alkylphenols and alkylphenol polyethoxylates. Water Qual Res J Can 34:123–177
Takeo M, Prabu SK, Kitamura C, Hirai M, Takahashi H, Kato D, Negoro S (2006) Characterization of alkylphenol degradation gene cluster in Pseudomonas putida MT4 and evidence of oxidation of alkylphenols and alkylcatechols with medium-length alkyl chain. J Biosci Bioeng 102:352–361. doi:10.1263/jbb.102.352
Tanghe T, Dhooge W, Verstraete W (1999) Isolation of a bacterial strain able to degrade branched nonylphenol. Appl Environ Microbiol 65:746–751
Tollefsen K-E, Eikvar S, Finne EF, Fogelberg O, Gregersen IK (2008) Estrogenicity of alkylphenols and alkylated non-phenolics in a rainbow trout (Oncorhynchus mykiss) primary hepatocyte culture. Ecotoxicol Environ Saf 71:370–383. doi:10.1016/j.ecoenv.2007.10.006
Toyama T, Sato Y, Inoue D, Sei K, Chang Y-C, Kikuchi S, Ike M (2009) Biodegradation of bisphenol A and bisphenol F in the rhizosphere sediment of Phragmites australis. J Biosci Bioeng 108:147–150. doi:10.1016/j.jbiosc.2009.03.011
van der Maarel MJEC, Kohler H-PE (1993) Degradation of 2-sec-butylphenol: 3-sec-butylcatechol, 2h-hydroxy-6-oxo-7-methylnona-2, 4-dienoic acid, and 2-methylbutyric acid as intermediates. Biodegradation 4:81–89
White R, Jobling S, Hoare SA, Sumpter JP, Parker MG (1994) Environmentally persistent alkylphenolic compounds are estrogenic. Endocrinology 135:175–182
Ying GG, Williams B, Kookana R (2002) Environmental fate of alkylphenols and alkylphenol ethoxylates—a review. Environ Int 28:215–226
Acknowledgments
This research was supported by a Grant-in-Aid for Young Scientists (B) (no. 19710060) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Toyama, T., Maeda, N., Murashita, M. et al. Isolation and characterization of a novel 2-sec-butylphenol-degrading bacterium Pseudomonas sp. strain MS-1. Biodegradation 21, 157–165 (2010). https://doi.org/10.1007/s10532-009-9290-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-009-9290-y