Abstract
Recommendations are a key component of radiology reports. Automatic extraction of recommendations would facilitate tasks such as recommendation tracking, quality improvement, and large-scale descriptive studies. Existing report-parsing systems are frequently limited to recommendations for follow-up imaging studies, operate at the sentence or document level rather than the individual recommendation level, and do not extract important contextualizing information. We present a neural network architecture capable of extracting fully contextualized recommendations from any type of radiology report. We identified six major “questions” necessary to capture the majority of context associated with a recommendation: recommendation, time period, reason, conditionality, strength, and negation. We developed a unified task representation by allowing questions to refer to answers to other questions. Our representation allows for a single system to perform named entity recognition (NER) and classification tasks. We annotated 2272 radiology reports from all specialties, imaging modalities, and multiple hospitals across our institution. We evaluated the performance of a long short-term memory (LSTM) architecture on the six-question task. The single-task LSTM model achieves a token-level performance of 89.2% at recommendation extraction, and token-level performances between 85 and 95% F1 on extracting modifying features. Our model extracts all types of recommendations, including follow-up imaging, tissue biopsies, and clinical correlation, and can operate in real time. It is feasible to extract complete contextualized recommendations of all types from arbitrary radiology reports. The approach is likely generalizable to other clinical entities referenced in radiology reports, such as radiologic findings or diagnoses.
Similar content being viewed by others
References
Mabotuwana T, Hall CS, Hombal V, Pai P, Raghavan UN, Regis S, et al: Automated tracking of follow-up imaging recommendations. AJR Am J Roentgenol. 2019;1–8.
Zafar HM, Chadalavada SC, Kahn CE Jr, Cook TS, Sloan CE, Lalevic D, et al: Code Abdomen: An Assessment Coding Scheme for Abdominal Imaging Findings Possibly Representing Cancer. J Am Coll Radiol. 2015;12: 947–950.
Steinkamp JM, Chambers C, Lalevic D, Zafar HM, Cook TS: Toward Complete Structured Information Extraction from Radiology Reports Using Machine Learning. J Digit Imaging. 2019;32: 554–564.
Dang PA, Kalra MK, Blake MA, Schultz TJ, Halpern EF, Dreyer KJ: Extraction of recommendation features in radiology with natural language processing: exploratory study. AJR Am J Roentgenol. 2008;191: 313–320.
Dang PA, Kalra MK, Blake MA, Schultz TJ, Stout M, Lemay PR, et al: Natural language processing using online analytic processing for assessing recommendations in radiology reports. J Am Coll Radiol. 2008;5: 197–204.
Dreyer KJ, Kalra MK, Maher MM, Hurier AM, Asfaw BA, Schultz T, et al: Application of recently developed computer algorithm for automatic classification of unstructured radiology reports: validation study. Radiology. 2005;234: 323–329.
Yetisgen-Yildiz M, Gunn ML, Xia F, Payne TH: A text processing pipeline to extract recommendations from radiology reports. J Biomed Inform. 2013;46: 354–362.
Mabotuwana T, Hall CS, Dalal S, Tieder J, Gunn ML: Extracting Follow-Up Recommendations and Associated Anatomy from Radiology Reports. Stud Health Technol Inform. 2017;245: 1090–1094.
Reddy S, Chen D, Manning CD: CoQA: A Conversational Question Answering Challenge. Transactions of the Association for Computational Linguistics. 2019;7: 249–266.
Park DH, Hendricks LA, Akata Z, Rohrbach A, Schiele B, Darrell T, et al: Multimodal explanations: justifying decisions and pointing to the evidence. arXiv [cs.AI]. 2018. Available: http://arxiv.org/abs/1802.08129
Sukthanker R, Poria S, Cambria E, Thirunavukarasu R: Anaphora and coreference resolution: a review. arXiv [cs.CL]. 2018. Available: http://arxiv.org/abs/1805.11824
12. Hochreiter S, Schmidhuber J: Long short-term memory. Neural Comput. 1997;9: 1735–1780.
[PDF]GloVe: Global vectors for word representation—Stanford NLP. Available: https://nlp.stanford.edu/pubs/glove.pdf
Segura Bedmar I, Martínez P, Herrero Zazo M: Semeval-2013 task 9: Extraction of drug-drug interactions from biomedical texts (ddiextraction 2013). Association for Computational Linguistics; 2013. Available: https://e-archivo.uc3m.es/bitstream/handle/10016/20455/semeval_SEMEVAL_2013.pdf?sequence=3
Zhang Y, Yang Q: A survey on multi-task learning. arXiv [cs.LG]. 2017. Available: http://arxiv.org/abs/1707.08114
Sistrom CL, Dreyer KJ, Dang PP, Weilburg JB, Boland GW, Rosenthal DI, et al: Recommendations for additional imaging in radiology reports: multifactorial analysis of 5.9 million examinations. Radiology. 2009;253: 453–461.
Cochon LR, Kapoor N, Carrodeguas E, Ip IK, Lacson R, Boland G, et al: Variation in Follow-up Imaging Recommendations in Radiology Reports: Patient, Modality, and Radiologist Predictors. Radiology. 2019;291: 700–707.
Kaissis GA, Makowski MR, Rückert D, Braren RF: Secure, Privacy-Preserving and Federated Machine Learning in Medical Imaging. Nature Machine Intelligence 2020;6: 305–11.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
JS is part-owner of River Records LLC., a healthcare company focused on developing tools to reduce clinician documentation burden and improve healthcare quality. The company had no involvement in this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix 1. Annotation Protocol for Specific Questions
Question 1: What are all of the Recommendations in this Report?
To fully capture the diversity of recommendations in reports, as well as to enable the maximal variety of future descriptive analyses or clinical tracking systems, we defined recommendations as broadly as possible. Recommendations were defined as any span of text indicating that something should or should not be done or considered by the ordering clinician. This includes recommendations for imaging studies, lab tests, tissue samples, specialty referrals, correlation with physical exam findings, comparison with prior imaging, and nonspecific statements of “clinical correlation.” Statements that “No further follow-up is required” are annotated as negated recommendations; i.e., “follow-up” is tagged as the text span answer to the top-level question, and “no” is the classification answer to the downstream question “Is this recommendation negated?” Statements that “XXX may be considered for better visualization” were included as recommendations.
Question 2: What is the Desired Time Period for this Recommendation?
Any statement indicating a time period, whether exact, relative, or vague, was annotated as a text-span answer to this question (e.g., “Screening mammogram in 12 months,” “short interval follow-up,” “nonemergent CT.” “MRI when patient’s clinical condition permits”).
Question 3: What are the Stated Reasons for this Recommendation?
Text spans were annotated as answers to Question 3 if they provided context for the recommendation regarding its purpose, reason, or goal. This includes broad indication categories (“screening mammogram,” “follow-up imaging,” “surveillance”), specific stated goals for follow-up (“to rule out malignancy,” “to ensure resolution of consolidation”), rationale or justification statements (“given stability over the past 12 months”), and findings which clearly motivated the recommendation (“'Indeterminate left adrenal mass may represent atypical adenoma and should be characterized. Recommend…”). For negated recommendations, reasons for not performing the follow-up were annotated. This was designed to be used in downstream systems which link recommendations to findings over time, with the understanding that it is somewhat more difficult than the other questions to define exactly.
Question 4: Under What Conditions Should this Recommendation be Performed?
Many recommendations include conditions which should be met before the recommendation is done (e.g., “if the patient is at increased risk for pulmonary metastatic disease,” “if further evaluation of this finding is desired,” “if clinically indicated,” “otherwise”). Any such condition was tagged as a free-text answer to this question.
Question 5: What is the Strength of this Recommendation?
While this can be a category of dispute even among radiologists, we opted to use a few simple heuristics for annotation. Statements such as “is recommended,” “is advised,” or imperative directives (“Follow up in 6 months”) were annotated as strong recommendations, whereas those which “suggested” asked ordering clinicians to “consider” a recommendation or posited that a follow-up study “may be useful” were annotated as weak recommendations. Similarly, recommendations which merely stated that a follow-up study has the potential to better distinguish differential diagnoses were annotated as weak recommendations.
Question 6: Is this Recommendation Explicitly Negated?
Many recommendations are actually statements not to do something, or that nothing is necessary to be done. If a recommendation was of this type (e.g., “No further follow-up is required,” “Does not meet criteria for follow-up”), the recommendation was annotated as negated, and the negation phrase (e.g., “no,” “nothing,” “does not”) was tagged as a free-text rationale for the classification.
Appendix 2. Custom Labeling Application
Our custom web application for report annotation is demonstrated below. During annotation time, it was hosted behind our institution’s firewall, accessible only on the institution’s internal network. The application was hosted using Python’s Flask web server software, with a MongoDB database for storing documents and annotations. The front-end client-side interface was written using Vue.js. The web application allowed for the creation of annotation projects, each of which has their own set of “questions.” Documents were loaded into the application and tokenized using SpaCy. The interface enables users to navigate between different questions and annotate documents at the token or character-level. In the case of this project, annotations were performed at the token level.
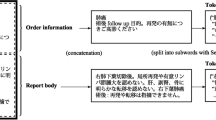
Figure 1 shows the web interface being used to annotate a sample anonymized radiology report.
(a) The custom web interface used to annotate reports. This shows a user annotating a top-level question (question 1) for an anonymized radiology report. The user uses the left-hand column to select the question to annotate answers for and uses the center column to highlight and tag spans of text as answers to that particular question. In this document, there are three separate recommendations, corresponding to three answers to the “recommendation” question. (b) In this screenshot, the user is annotating question 2 (“What is the desired time period for this recommendation?”) for each of the three previously identified recommendations in this report. The right-hand column is used to select which follow-up recommendation (i.e. which answer to question 1) to annotate. Note that downstream questions such as question 2 may also have multiple answers, as in this case –“CT” should be performed “annually” and “for 5 years.” Both of these text spans provide temporal context for the recommendation
Appendix 3. Neural Network Architectures
Word Vectors
In order to capture the specifics of the relatively limited domain of radiologic text while also maintaining the benefits of word embeddings trained on a large and diverse English corpus, we opted to combine two word embeddings: (1) a set of general-purpose English text embeddings from spaCy’s “en_core_web_lg” model (trained on the Common Crawl dataset as Global Vectors (GloVe)) and (2) custom-trained fastText vectors on our institution’s entire corpus of radiology reports. We used the fastText implementation from the gensim python package with the skip-gram training procedure, 300-dimensional embeddings, and all other parameters set to gensim’s default values.
LSTM Multi-Task Model
The multi-task LSTM consisted of a document embedding layer with an output consisting of 600 dimensions; 300 of these came from the default SpaCy vectors (‘en_core_web_lg’ model), and 300 from our custom-trained fastText vectors. In addition, there was a separate referent embedding layer which embedded the referent vector in a 300-dimensional space. For this study, there was only one type of referent (recommendations), so the embedding layer processed vectors consisting entirely of zeros (indicating that the token was not part of a referent) and ones (indicating the token was part of a recommendation referent), although it is generalizable to an arbitrary number of referent types. These two word embeddings were concatenated to produce the final token embedding of size 900. The concatenated embeddings were then processed by 2 bidirectional LSTM layers of dimension 400. The processed text was concatenated, token-wise, with the question embedding. The question embedding was a third, separate embedding layer which embedded the question type, enabling the same network to answer all six different questions. A two-layer dense neural network (dimensions: 700 × 350 followed by 350 × 2), with an intermediate ReLU layer to allow for nonlinearity, was then used to process each token to produce the final two-dimensional output (part of an answer vs. not). For questions with categorical inputs, a maxpool over all tokens was applied before a separate 2-layer dense network (700 × 350, 350 × 2). The network is shown in Fig. 2a.
(a) Diagram of the multi-task neural network model. Green boxes represent model actions; blue boxes represent the state of data as it passes through the network. |V|: number of tokens in vocabulary. |Q|: number of unique questions to be answered by the network. L: length of document. Batch sizes are ignored for ease of comprehension. (b) Diagram of the single-task neural network model. Green boxes represent model actions; blue boxes represent the state of data as it passes through the network. In this model, the question type is used to select which of the sub-networks handles the question, and therefore requires no question embedding required. Each sub-network has its own unique weights and is trained only on a single question. |V|: number of tokens in vocabulary. |Q|: number of unique questions to be answered by the network. L: length of document. Batch sizes are ignored for ease of comprehension
LSTM Single-Task Model
This model is very similar to the multi-task model; the only difference is that there is a separate network (with unique weights) for each question, which only answers that question. The type of question now merely determines which sub-network each question is passed to. The network is shown in Fig. 2b.
Rights and permissions
About this article
Cite this article
Steinkamp, J., Chambers, C., Lalevic, D. et al. Automatic Fully-Contextualized Recommendation Extraction from Radiology Reports. J Digit Imaging 34, 374–384 (2021). https://doi.org/10.1007/s10278-021-00423-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-021-00423-8