Abstract
The broad objective of this research was to determine the environmental drivers of macroinvertebrate and microbial assemblages in acidic pit lakes. This is important because pit lake ecosystem development is influenced by prevailing environmental characteristics. Three lakes (Stockton, Kepwari, WO5H) within a larger pit-lake district in Collie, Western Australia were surveyed for spatial variability of benthic macroinvertebrate and microbe (Archaea, Bacteria) assemblage composition as well as potential environmental drivers (riparian condition, aquatic habitat, sediments, and aquatic chemistry) of assemblages. With the exception of sediment chemistry, biophysical variables were significantly different across lakes and reflected riparian condition and groundwater chemistry. Microbial assemblages in pit lakes were significantly different across lakes and correlated with water chemistry, particularly metals in Lake WO5H. However, the most abundant microbes were not readily identified beyond class, making it difficult to speculate on their ecological function. Macroinvertebrate assemblage composition and species richness were also significantly different across all lakes, and in Lake WO5H (a lake with low pH and high metal concentrations), taxa were correlated with benthic organic matter as well as water chemistry. Results indicated that despite poor water quality, input of nutrients from terrestrial leaf litter can support or augment pit lake ecosystems. This is a demonstration of the concept that connection of pit lakes to catchments can positively affect aquatic ecosystems, which can inform management actions for remediation.
Similar content being viewed by others
Introduction
Large-scale mining permanently changes landscapes. Mountaintop removal, construction of new landforms, and the creation of thousands of open pits occurs across every inhabited continent on earth (Blanchette and Lund 2016). Pit lakes form when these open pits fill with ground, surface and rain water, and by virtue of their size and water toxicity, can have substantial environmental impact (Lund and Blanchette 2018; Miller et al. 1996; Younger and Wolkersdorfer 2004). Historically, pit lake rehabilitation has been considered relatively unimportant, with lakes locked away from public access, either mandated for ‘perpetual treatment’ or simply abandoned with minimal (if any) safety measures (Blanchette and Lund 2016). There is growing recognition that successful mine closure is a stakeholder-driven process (Bainton and Holcombe 2018), and communities are increasingly unwilling to tolerate abandoned pit lakes on their doorstep (see Kean 2009; Woodbury 1998).
By virtue of their location, size, and geology, pit lakes can be considered to inhabit a point along a ‘sliding scale’ of interacting factors that increases the complexity of rehabilitation (Blanchette and Lund 2016). For example, a large lake with highly toxic water and prone to frequent tropical cyclones may lay at the more ‘difficult’ end of the scale. However, not all pit lakes are alike. Shallow (1–5 m) coal-strip lakes with large amounts of terrestrial leaf litter (Campbell and Lind 1969; Coe and Schmelz 1972) are now recreational fisheries in the US Midwest. The well-forested pit lake district of Lusatia, Germany has been developed for public recreational activities such as boating, upmarket hotels, spas, and restaurants, and is projected to contribute at least €10 M to the local economy (Lienhoop and Messner 2009). If these new lakes are to be community assets, they must be properly monitored and understood—beyond simple water quality measures—because community members expect water bodies to be safe, accessible, and biodiverse (sensu Naiman 2013).
Aquatic macroinvertebrates are routinely used to assess the condition of bioindicators of freshwater ecosystems. The ecology of aquatic macroinvertebrates is comparatively well-known, with an extensive body of literature to assist monitoring programs. Advances in technology can enhance monitoring programs by using DNA sequencing to identify macroinvertebrates (Carew et al. 2013) or measure gene expression to determine environmental stress (Chou et al. 2018). Increasingly, microbial assemblages (16S) are being used either alone or in conjunction with macroinvertebrates to characterise freshwater ecosystem condition and response to anthropogenic pressures (e.g. Horton et al. 2019; Simonin et al. 2019; Vander Vorste et al. 2019).
Relative to other inland waterbodies, the ecology of pit lakes is understudied and underrepresented in the primary literature (Blanchette and Lund 2016). As products of the mining industry, pit lake microbial ecology is readily viewed in terms of microbially-mediated biogeochemistry, and understanding these reactions can be used to improve water quality modelling (Wielinga 2009). Past studies used culture-dependent methods or cell counts to characterise communities, inadvertently missing much of the taxonomic diversity. More recent pit lake studies have adopted culture-independent 16S sequencing, which has expanded knowledge of mine pit lake microbial biodiversity (see González-Toril et al. 2015; Kampe et al. 2010; Lucheta et al. 2013), but few studies have compared the drivers of microbes and macroinvertebrates in pit lakes, or applied this knowledge to closure planning.
The broad objective of this research was to determine the drivers of macroinvertebrate and microbial assemblages in acidic pit lakes. This is important because pit lake ecosystem development is influenced by prevailing environmental characteristics (e.g. riparian condition, aquatic chemistry), which inform remediation, monitoring, and closure. Microbes are key to pit lake ecosystems; DNA sequencing technology has become more routine, but collection and analysis is still highly variable and there are currently no standard methods. This work will compare the environmental drivers of microbial assemblages (a relatively new bioindicator) with aquatic macroinvertebrates (an established bioindicator).
The specific aims of the research were to: (1) characterise the biophysical variables of three co-occurring mine pit lakes in Collie, Western Australia, (2) collect macroinvertebrates and microbes (Archaea, Bacteria) from the pit lakes, (3) determine if assemblages were significantly different among lakes, (4) seek biophysical variables that correlated with assemblages, and (5) ascertain if, in relation to macroinvertebrates, microbial assemblages provide alternative or complementary understanding of pit lake ecosystems.
Methods
Site Description
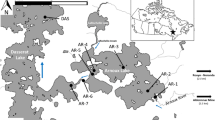
The three study lakes (Kepwari, Stockton, and WO5H) are former coal pits located in the Collie basin, Western Australia (Fig. 1). The coalfield itself is approximately 224 km2 with Permian sedimentary succession up to 1400 m thick—the top 900 m containing coal seams between 0.5 and 13 m thick (Millar et al. 2011). Mining began by hand in 1892; the first open cut operation started in 1943 (Millar et al. 2011), eventually creating a ‘district’ of 10 pit lakes (Fig. 1) variably hydrologically connected to the wider catchment by seasonal surface and ground water flows. Collie has a Mediterranean climate with dry summers (14–30 °C) and wet, cold winters (4–16 °C) (mean rainfall for years 2002–2019 was 711 mm; May–Sept was 539 mm; Australian Bureau of Meteorology 2019).
Lake Stockton (Fig. 2) was opened to underground mining in 1927 and open-cut operations began in 1943, with both types of mining active until it was abandoned in 1960 (Stedman 1988). After abandonment, the pit filled naturally with groundwater, creating a lake 25–47 m deep that is currently used for public recreation, particularly water skiing. Lake Stockton has occasionally received dewatering water from Griffin Coal operations, and the lake water periodically discharges into a creek that meets the Collie River South (see Lund and Blanchette 2018).
Pit WO5H (Fig. 2) was mined from 1986 to 1997 and then rapidly filled with surface water until 2004 (McCullough et al. 2009). An acidic (pH 4.9), saline stream flows into Lake WO5H due to seasonal rainfall events, and the lake can discharge, with lake water joining flows from Lake Stockton before running into the Collie River South (McCullough et al. 2009). Discharge from WO5H is prevented in most years by pumping substantial volumes of lake water for use in power station cooling. Efforts were made to plant trees and shrubs below the final water line before filling to add carbon to the highly oligotrophic lake (Lund et al. 2014).
The Collie River South was diverted around the WO5B pit to allow mining. Mining in the WO5B pit ceased in 1996, and it was rapidly filled from 1999 to 2004 from the Collie River South to form Lake Kepwari (Fig. 2), which is 65 m deep and covers an area of 1.03 km2 (McCullough et al. 2009). The overburden dumps have been extensively revegetated and the final intended use of the lake is as a recreational resource. The original closure plan was to keep the lake isolated from the Collie River South using the diversion; however, a large rain event in 2011 caused the river to breach the diversion and enter the lake, temporarily improving lake water quality by increasing the pH. The company was granted approval to connect the pit lake to the river permanently as part of the closure plan, which will see the lake relinquished back to the state of Western Australia (Lund and Blanchette 2018).
Field and Laboratory Protocols
Lakes WO5H, Stockton, and Kepwari were each sampled once in April 2013 (see “Data Analysis” section, below for experimental design). Macroinvertebrates and habitat features were collected from the littoral zone approximately every 200 m around the perimeter of each lake in an attempt to equalise sampling intensity among the lakes (Stockton n = 8, Kepwari n = 21, WO5H n = 14; perimeter distance determined using Google Earth © mapping tool). A surface water sample was collected from the centre of each lake for chemical analysis. Sediment for microbial and chemical analysis was collected from five sites at 2–3 m depth in each lake ≈ 5 m from the littoral zone (varying 1–2 m depending on prevailing conditions and safety for SCUBA divers). In situ measurements of surface physico-chemistry were collected at each site.
Macroinvertebrates were collected from a 0.5 m2 quadrat via vigorous sweep sampling for 20 s using a square dip net with 500 µm mesh and immediately preserved in 70% ethanol. Macroinvertebrate identifications (to species, where possible) were completed according to Williams (1980), Davis and Christidis (1997), Hawking and Theischinger (1999), Suter (1999), Watts (2002), and Gooderham and Tsyrlin (2002). In this study, zooplankton captured in the quadrat were counted as macroinvertebrates as they can compose the bulk of pit lake biota (Gammons et al. 2009).
In situ measurements of physico-chemistry (turbidity (NTU), chlorophyll a (µg L−1), pH, dissolved oxygen (DO; mg L−1), % saturation, conductivity (EC; mS cm−1), temperature (°C), and oxidation reduction potential (ORP; mV) were recorded using the Hydrolab DataSonde 4A (Hach Co., Loveland, CO, USA) ≈ 10 cm above the benthos (5–10 cm below the surface for macroinvertebrate transects and 2–3 m below the surface at sediment collection sites).
Water samples for metal and nutrient analysis were collected ≈ 10 cm below the surface of each lake. Each sample was split into three aliquots: one unfiltered and two filtered. Samples (where appropriate) were filtered through 0.5 µm Pall Metrigard™ filter paper under vacuum pressure. The unfiltered aliquot was frozen (− 12 °C) for later determination of total nitrogen (TN) and phosphorus (TP) following persulphate digestion as per APHA (1998a). One filtered aliquot was frozen (− 12 °C) for later determination of chloride (Cl−) using ion chromatography (Methrohm, Switzerland), nitrate/nitrite (NOx-N), filterable reactive phosphorus (FRP-P), and ammonia (NH3-N) on a Lachat (Hach, USA) autoanalyser (APHA 1998b), and dissolved organic carbon (DOC; measured as non-purgeable organic carbon) using a total carbon analyser (Shimadzu, Japan). The second filtered aliquot was acidified with nitric acid (to a pH < 2 ≈ 1% v/v) and then kept at 4 °C for later determination by ICP-AES/MS of a range of metals/metalloids (Al, As, B, Ca, Cd, Cr, Co, Cu, Fe, Hg, K, Mg, Mn, Na, Ni, Pb, Se and Zn) and S (APHA 1998b).
A 20 m riparian transect was established parallel to each macroinvertebrate collection site to record percent bank cover of grass, leaf litter cover, twigs/branches, exposed soil, canopy cover, standing dead trees, rushes, and shrubs as well as the extent of undercutting, slumping, gullying, and bank slope. Benthic cover was also quantified, including periphyton/algal mat, aquatic and semi aquatic macrophytes, leaf litter, and coarse particulate organic matter (CPOM). A Krumbein phi-scale was used to categorise sediment composition with local modifications for data analysis (sediment size index 1: sand-dominated, few rocks > 0.5 cm diameter (d), 2: silt-dominated, few rocks > 0.5 cm d, 3: sand and silt, few rocks > 0.5 cm d, 4: complex rocky mix—little sand/silt, rocks < 25 cm d, 5: boulders present > 25 cm d (Blanchette and Lund 2017; Blanchette and Pearson 2012, 2013).
Sediments were collected by SCUBA divers using modified gamma-sterile syringes, ensuring that only the top 5 cm was retained for microbial and chemical analysis (from Blanchette et al. 2019). Samples were immediately put on ice for transport, kept at − 20 °C for short-term field storage, and finally − 80 °C in the lab. Benthic sediments (7 to − 3 on phi scale) were analysed for bioavailable metals and nutrients according to U.S. EPA protocol 3050B (USEPA 1996) at the Edith Cowan University Analytical Chemical Laboratory using the Thermo Fisher Scientific iCAP QICPMS and 7600 ICPOES (Blanchette et al. 2019). Total N (Rayment and Lyons Method 7A5) and total organic C (acidification method 6B1) (Rayment and Lyons 2011) were analysed at CSBP Perth, Australia.
Microbiome Analysis
DNA was extracted from 750 µg of wet sediment using the FavorPrep™ Soil DNA Isolation Mini Kit and quantified using a Qubit® 3.0 Fluorometer (Thermo Fisher Scientific Inc.) according to manufacturers’ protocols. Microbiome analysis (16S rRNA sequencing) was performed by the Lotterywest State Biomedical Facility Genomics at the University of Western Australia according to Caporaso et al. (2012) with local modifications and detailed in Nagel et al. (2016).
Samples (1 ng) were amplified using the 16S V4/5 primers (515F: GTGCCAGCMGCCGCGGTAA and 806R: GGACTACHVGGGTWTCTAAT). To minimise primer–dimer formation and streamline downstream purification we used a mixture of gene-specific primers and gene-specific primers tagged with Ion Torrent-specific sequencing adaptors and barcodes (ratio of tagged and untagged primers was 90:10). Using this method, the ≈ 10-cycle inhibition observed by using long-tagged primers could be reversed and all samples were amplified using 18–20 cycles (5PRIME HotMasterMix, 5PRIME, USA) and confirmed by agarose gel electrophoresis. PCR product formation was quantified with a Qubit, as above. A maximum of 100 samples were diluted to equal concentrations and adjusted to a final concentration of 15 pM. Templated ion sphere particles were generated using an Ion Chef (Thermo Fisher Scientific) with a 400 bp templating kit and sequenced on an Ion PGM (Thermo Fisher Scientific) (850 cycles, 400 bp sequencing kit, modal read length of 309 bp). Reads were trimmed to improve quality using Torrent Suite 5.0 (Thermo Fisher Scientific). Taxonomic names were assigned to operational taxonomic units (OTUs) using software analysis programme Quantitative Insights into Microbial Ecology (QIIME, version 1.7) (Caporaso et al. 2010). OTUs based on 97% similar specific 16S rRNA sequence identities were used to distinguish different ‘species’ (or OTUs) of microbes using Greengenes database (McDonald et al. 2012; version 12_10). Samples were rarefied at 5000 reads for calculating dissimilarity between samples using UniFrac metrics.
Data Analysis
The overall experimental design incorporated collection of samples for aquatic macroinvertebrates and benthic microbes (Archaea, Bacteria) from three lakes during April 2013. To facilitate interpretation, biophysical and chemical data were separated into different matrices for analysis: (1) biotic (aquatic habitat and riparian characteristics), (2) water quality (physico-chemical and water chemistry), and (3) sediment chemistry (sensu Blanchette and Lund 2017). Biotic and in situ physico-chemical data were collected at each macroinvertebrate transect, and in situ physico-chemical data and sediment chemistry data at each microbe collection site. We tested the null hypotheses of no significant difference in water quality variables among lakes and no significant differences in macroinvertebrate or microbe assemblages among lakes. Biotic, water quality, sediment chemistry, and organism data were then ordinated to illustrate multivariate patterns and correlated against ordination axes to elucidate highly-correlated taxa. Correlations between biotic, water quality, and sediment chemistry variables and organism data were first tested for significance before relationships with specific parameters were explored.
Biotic, water quality, and sediment chemistry were compared among lakes using PERMANOVA in PRIMER-e with the Euclidean distance measure on normalised data (Anderson et al. 2008). Lake was treated as a fixed factor and transect was a random factor nested within lake (9999 permutations, significant p < 0.05), and pseudo-F is reported as F (Anderson et al. 2008). Biotic, water quality, and sediment chemistry data were ordinated with principal components analyses (PCA) in PRIMER-e. Data were predominantly linearly related to ordination axes and the significance of this relationship was assessed using Pearson’s r (α = 0.01) (Blanchette and Pearson 2012).
While 43 macroinvertebrate samples were collected from all three lakes (as above), two transects (one each from Stockton and WO5H) contained no animals and were thus excluded from analysis (41 samples in final analysis; Stockton n = 7, Kepwari n = 21, WO5H n = 13). Rare macroinvertebrate taxa (< 0.01%) were excluded to decrease noise in the data set and improve detection of correlations between assemblages and biophysical variables (Blanchette and Pearson 2012; Cao et al. 2001; Herlihy et al. 2005; Marchant 2002), reducing an initial count of 40 taxa (across all lakes) to 19. The results therefore only describe common taxa. Macroinvertebrate species richness (S; common taxa only) was determined as mean S per quadrat per lake, and was calculated using one-way ANOVA in SigmaPlot 13.0 with a Tukey pairwise test (Systat Software, Inc.). Abundances were log (x)-transformed prior to analysis (Anderson et al. 2008; Blanchette and Pearson 2012). Macroinvertebrate log(abundance) data, hereafter referred to as ‘assemblage data’, were compared among lakes using PERMANOVA in PRIMER with the Bray–Curtis distance measure; again, lake was treated as a fixed factor and transect was a random factor nested within lake (9999 permutations, significant p < 0.05) and pseudo-F is reported as F (Anderson et al. 2008). Macroinvertebrate assemblage data were ordinated with nonmetric multidimensional scaling (NMDS) in PRIMER-e—a 2- or 3-dimensional solution was optimal for all runs (stress below 0.20). Pearson–Kendall correlation (Pearson’s r, as per biophysical variables, above) was used to identify relationships between macroinvertebrate assemblage data with NMDS axes (Blanchette and Pearson 2012; Herlihy et al. 2005). Correlations between macroinvertebrate assemblage data and biotic, water quality and sediment chemistry data were analysed using RELATE (Spearman’s rank; rs, p < 0.05; 9999 permutations) with a Pearson’s correlation in PRIMER-e.
While 15 sediment samples were collected from all three lakes (as above), two transects (one each in Stockton and WO5H) did not contain sufficient DNA to amplify for downstream application and were thus excluded from analysis (13 samples in final analysis; Stockton n = 4, Kepwari n = 5, WO5H n = 4). Hypothesis tests on OTU (operational taxonomic unit) weighted (abundance and presence/absence) UniFrac genetic distance data were conducted with a PERMANOVA in PRIMER-e and ordinated with NMDS (factors, permutations and stress as per macroinvertebrate data). Correlations between OTU data and sediment chemistry and water quality were analysed using RELATE as per macroinvertebrate assemblage data and Blanchette et al. (2019).
Results
Biophysical Variables
Aquatic and riparian characteristics (biotic parameters; Supplementary Table 1) were significantly different across lakes (PERMANOVA; F(2, 38) = 3.96, p < 0.001), and biotic parameters at all lakes were significantly different from each other (PERMANOVA pair-wise; all lake comparisons p < 0.001). A PCA plot (Axis 1; 18.6% of the variance, Axis 2; 16.2%, Axis 3; 12.8%) separated Lake Stockton from lakes Kepwari and WO5H along Axes 1 and 2 (Fig. 3). Pearson–Kendall correlation analysis (|r| = 0.01, 40 ≥ 0.393) indicated that PCA axis 1 was most strongly correlated with bank geomorphology (slumping, undercutting, slope; |r| > 0.700), PCA axis 2 was most strongly correlated with vegetation (rushes, riparian tree condition; |r| > 0.670), and PCA axis 3 was most strongly correlated with leaf litter (|r| > 0.629). Relative to Lakes Kepwari and WO5H, Stockton had potentially more bank instability, a greater cover of aquatic plants/rushes, and less terrestrial leaf litter input (Supplementary Table 1).
In situ water quality (Table 1) and water chemistry (Table 2) data were significantly different across lakes (PERMANOVA; F(2, 38) = 154.47, p < 0.001), as were pairwise comparisons of these data between all lakes (p < 0.001). A PCA plot (Axis 1; 69.9% of the variance, Axis 2; 19.6%, Axis 3; 4.8%) separated all lakes along axis 1, whereas Lake Stockton was separated from lakes Kepwari and WO5H along axis 2 (Fig. 3). Pearson–Kendall correlation analysis (|r| = 0.01, 40 ≥ 0.393) indicated that PCA axis 1 was most strongly correlated with metal concentration, sulphur, ammonium, and pH [e.g. Ni, Al, Fe, Mn, Zn, pH, sulphur, NH4+|r| > 0.940) and axis 2 was correlated with Ca (|r| = 0.940)]. Lake WO5H had higher concentrations of metals, sulphur, and ammonium, and a lower pH than lakes Stockton and Kepwari. Lake Stockton had the lowest Ca concentration (Table 1, Supplementary Table 2).
Sediment chemistry data were not significantly different across lakes (PERMANOVA; F(2, 10) = 1.96, p = 0.066) (Table 3).
Macroinvertebrates
In total, 81,466 individuals from 19 aquatic taxa were identified across all lakes (Supplementary Table 2). Planktivores composed 98.5% of the individuals collected, which were mainly calanoid copepods (98.4%) from lakes Kepwari and Stockton. Calanoid copepods were not collected from WO5H. The detriti-herbivore Chironomini (Chironominae) was the most abundant insect taxon (1.80%), with high abundances in Lake WO5H and few individuals found in lakes Kepwari and Stockton. Predators were rare (1.28%), but also were the most diverse group, even at high taxonomic levels within the Insecta (Diptera, Odonata, Coleoptera) as well as the Arachnida (Acarina; Orabatidae). Species richness (S) was significantly different among lakes (F(2,40) = 8.76, p < 0.001), and results of a Tukey test indicated that mean species richness per quadrat between lakes Kepwari and Stockton (p < 0.001) and WO5H and Stockton (p = 0.034) were significantly different (S ± SE: Kepwari; 7.29 ± 0.55, Stockton; 3.25 ± 0.77, W05H; 5.93 ± 0.56).
Overall, macroinvertebrate assemblages were significantly different among lakes (PERMANOVA; F(2, 38) = 26.35, p < 0.001), and assemblages from all lakes were significantly different from each other (PERMANOVA pairwise; all lakes p < 0.001). An NMDS ordination of lake macroinvertebrates (2-dimensional solution, stress = 0.09) showed strong separation of Lake WO5H from lakes Kepwari and Stockton (Fig. 4). Pearson–Kendall correlation analysis (|r| = 0.01, 40 ≥ 0.393) indicated that, although relatively rare, predators and omnivores tended to have high r-values relative to NMDS axes and were associated with Lake WO5H (NMDS axis 1; Orthetrum caledonicum (Libellulidae), Chironomini (Chironominae), Necterosoma darwini (Dytiscidae), Ecnomina sp. (Trichoptera), |r| > 0.500, NMDS axis 2; Ceratopogoninae (Ceratopogonidae), |r| = 0.547).
Results of a RELATE procedure for macroinvertebrate assemblages and aquatic habitat and riparian characteristics indicated that the relationship was significant (rs = 0.160, p = 0.018). Macroinvertebrate assemblages on NMDS axis 1 were correlated with CPOM and benthic leaf litter cover (|r| > 0.500) in Lake WO5H, and aquatic plants (|r| = 0.394, NMDS axis 2) in Lake Stockton. There was also a significant relationship between macroinvertebrate assemblages and water quality (RELATE; rs = 0.728, p < 0.01). On NMDS axis 1, assemblages in Lake WO5H were correlated with higher concentrations of metals (Cd, Ni, Co, Al, Cu, Cr, Fe, |r| > 0.931) and lower pH (|r| = 0.932), whereas on NMDS axis 2 assemblages in Lake Stockton were correlated with lower Ca concentrations and lower conductivity measurements (|r| > 0.460).
Microbes
Microbes (UniFrac distance) were significantly different among lakes (PERMANOVA; F(2, 10) = 2.38, p < 0.001), and assemblages from all lakes were significantly different from each other (all pairwise comparisons were p < 0.05). Similar to the lake macroinvertebrates, an NMDS ordination of lake microbes (2-dimensional solution, stress = 0.11) showed separation of Lake WO5H from lakes Kepwari and Stockton (Fig. 5).
Across all lakes, there were 298 unique OTU categories from 178 families of microbes, seven of which were from kingdom Archaea (most were bacteria). Most microbes were rare; 281 of the 298 OTUs had average relative abundances (RAs) of less than 0.01, 17 OTUs had an average RA of 0.01–0.06, and one OTU had an average RA of 0.30, which was an unidentified bacterium (Fig. 6). Of the top nine OTUs by RA (Fig. 6), only seven of the taxa could be identified to class and one could be identified to family. OTU (species) richness (S) was significantly different among lakes (F(2, 10) = 5.40, p = 0.026), which was driven by a significant difference in mean S per sample between lakes Kepwari and WO5H (Tukey’s test; p = 0.024, S ± SE: Kepwari; 139.4 ± 23.9, Stockton; 112.8 ± 18.0, W05H; 100.0 ± 7.39).
Results of a RELATE procedure for microbes (UniFrac distance) and aquatic chemistry (in situ and analyses) indicated that the relationship was significant (rs = 0.456, p < 0.001). Similar to the macroinvertebrates, microbes in Lake WO5H were correlated (Pearson’s r; |r| = 0.01, 13 ≥ 0.641) with higher metal concentrations (NMDS axis 2; Cu, Cd, Ni, Co, Al, |r| > 0.860) and lower pH (|r| = 0.870). Unlike the macroinvertebrates, Ca and in situ conductivity were not significantly correlated with NMDS axes. Instead, turbidity was the only significantly correlated water quality variable associated with NMDS axis 1 (|r| = 0.840), separating Lake Stockton from lakes Kepwari and WO5H (n.b., although partially driven by a highly turbid data point in Lake Stockton). The relationship between microbes (UniFrac distance) and sediment chemistry was not significant (RELATE; rs = − 0.016, p = 0.47).
Discussion
The broad aim of this research was to compare the environmental drivers of a routinely-used bio-assessment tool (macroinvertebrates) with those of microbial assemblages (Archaea, Bacteria) in acidic pit lakes. The purpose of this comparison was to determine if microbes, which are rarely used in freshwater bio-assessment programs, provide complementary or alternative understanding of pit lake ecosystems. We found that in many ways, the two groups were complementary; their assemblages were significantly different across lakes and correlated with similar water quality and biotic characteristics, particularly the metals in Lake WO5H. This is an encouraging step towards developing a standardised collection protocol for benthic microbes—critical for progressing any new monitoring program. However, we were unable to identify many of the microbes, and in this respect, the macroinvertebrates provided more information regarding ecosystem processes. Given the extensive body of literature regarding the identification and ecology of macroinvertebrates, this research indicated that aquatic macroinvertebrates would be more useful in pit lake monitoring programs than microbes. Nevertheless, rapid advances in technology and decreases in per sample costs suggest that microbes as environmental bio-indicators is a line of enquiry worth pursuing.
Biophysical variables were significantly different across lakes, which reflected variable riparian conditions and groundwater characteristics. Lakes Kepwari and WO5H had more input of terrestrial leaf litter and more gently sloping banks than Lake Stockton, where signs of bank instability were more evident (i.e. slumping, undercutting, gullying). Bank instability in Lake Stockton may have been partially due to erosive wave action from recreational water skiing. Relative to other lakes, Stockton had a greater cover of true aquatic plants, but cover of both erosion and plants was patchy, and plants occurred in areas of limited erosion. Metal concentrations (e.g. Al, Cd, Cr, Ni, Zn) in Lake WO5H were high, likely because most of the drainage from the adjacent mine site collects in the lake were from surface run-off or interflows. Aluminium concentrations were particularly high at 21.7 mg L−1, although Cd, Cr, Ni and Zn exceeded the ANZECC/ARMCANZ (2000) guideline values for the 95% protection of aquatic ecosystems by up to 300%.
Macroinvertebrate assemblage composition and species richness was significantly different across all lakes, despite the close proximity of lakes within the ‘lake district’ and the prevalence of hardy, cosmopolitan taxa in the region (Smith et al. 1999). Across all lakes, predators were rare, but diverse, and calanoid copepods were the most relatively abundant organism collected (at 98.4%), but were not found in Lake WO5H. Planktonic communities may dominate pit lake biota, and their prevalence depends heavily on water quality (see Gammons et al. 2009). Lake WO5H was distinct from the other two study lakes by having high metal concentrations, a well-known inhibitor of zooplankton establishment in mine-affected oligotrophic lakes (Roch et al. 1985). Even relatively low metal concentrations in acidic and/or saline mine water can be deleterious to zooplankton, either directly or by compromising phytoplankton food items (Leppänen et al. 2017). While this was not a food web study per se, the total absence of calanoid copepods (the most dominant taxon of the study) at Lake WO5H indicates that the invertebrate-planktonic food web in WO5H was different from those of Lakes Kepwari and Stockton.
Relative to the other study lakes, Lake WO5H contained more predators and detriti-herbivores, which were significantly correlated with CPOM and benthic leaf litter cover. Lake WO5H also had a mean species richness nearly double that of Lake Stockton (but less than Lake Kepwari). These results suggest that despite poor water quality indicators of low pH and high metal concentrations, nutrient input from terrestrial leaf litter can support or augment pit lake ecosystems (sensu Blanchette and Lund 2016). After post-abandonment pit contouring, Lake WO5H underwent a planting program where native terrestrial seed was introduced to the pit below the final water level, allowing plants to be flooded as the pit filled (see Lund et al. 2014). Input of terrestrial leaf litter—as habitat and food—is a critical function of riparian zones, and riparian structure appears to be the main factor influencing leaf litter input (Naiman and Decamps 1997). Mine pit lake catchments have traditionally been considered risks due to acid mine drainage, public safety, and discharge (McCullough and Lund 2006). This research indicates that pit lake riparian zones can also provide an opportunity to enhance aquatic biodiversity, despite potentially poor water quality. While this study didn’t test if organisms were responding to terrestrial leaf litter as either ‘habitat’ or ‘food’ (e.g., Karádi-Kovács et al. 2015; Richardson 1992), future research will employ techniques such as stable isotope analysis to determine basal food web sources and relationships among taxa.
Despite differences in aquatic chemistry, measured sediment chemistry was not different among lakes. Compared to water quality variables, sediment chemistry was extremely spatially patchy within lakes and our small sample size (necessitated by cost) may not have fully captured within-lake variability. In an acidic Ontario pit lake, metal accumulations in the sediments were also highly spatially variable, and likely due to localised discharges of groundwater upwelling—discharge locations were ‘encrusted’ with iron and co-precipitated metals occurred directly next to sediments with low metal concentrations (Kalin and Wheeler 2013). Lake sediments are considered a “record of lake history” (Jones and Bowser 1978), and even in these apparently simple systems, depositional and hydrological processes produce a complicated benthic system. Future work will attempt to elucidate how variables such as sedimentation and mineralisation affect the within-lake spatial variability of sediments.
We took a classic ecological approach to the microbial study: collect organisms and biophysical variables, then statistically analyse and determine any correlations in the data. This approach worked well in seasonal rivers in New South Wales (Blanchette et al. 2019). However, in pit lakes sediment, microbial assemblages were not correlated with sediment chemistry, but rather water quality. One possibility is that we did not target the variables that actually drove differences in pit lake microbial assemblages—such as the precise nutrient fraction. Another possibility is that the microbes we collected were predominantly from the pelagic fraction of the food web, rather than the sediments. The benthos of the study lakes were mainly composed of kaolinite clay, which are powdery and mobile. It would be difficult for a distinct benthic biofilm community to form on a mobile sediment with little integrity (Gerbersdorf and Wieprecht 2015). Interstitial pore water and handling error by divers could have also introduced pelagic DNA to samples. Essentially, if the bottom of a lake is considered a ‘depositional reservoir’ (sensu Jones and Bowser 1978), we may have sequenced the DNA of dead/dying and/or transient pelagic microbes, rather than a true benthic community. Future work will collect pelagic microbes at different depths as well as ‘benthic’ microbes (as in Blanchette et al. 2019) and identify assemblages from sediment cores at different depths.
As discussed above, microbial assemblages in pit lakes were correlated with water chemistry, particularly metals and low pH, which separated Lake WO5H (which had the lowest OTU richness) from lakes Kepwari and Stockton. Microbial assemblages were also correlated with high turbidity in Lake Stockton. Many taxa were unable to be identified beyond class, and the most abundant taxon was an unidentified bacterium. However, in a September 2014 study of microbial assemblages in the Collie River South Branch (which flows through Lake Kepwari—see Lund et al. 2018), using the methods above, we (the authors) described 1239 OTUs from 591 different microbial families (Bacteria and Archaea; 428 ± 126 OTU richness per sample), most of which could be identified to genus (Lund and Blanchette 2018). In the river, bacteria unidentified at the phylum level were extremely rare in samples. Data from the river demonstrate that DNA techniques and libraries were sufficient to sequence and identify benthic aquatic microbes in the region.
Relative to co-occurring natural waterbodies, microbial assemblages in these acidic pit lakes were depauperate, and many did not occur in DNA databases at the time of analysis. While it is challenging to compare OTU richness among studies due to the potential effects of methodology, acidic pit lakes in Collie have similar OTU richness (100–130 per sample) to environments such as sea ice (128 OTU richness per sample; Yergeau et al. 2017), and hot springs (100–300 per-sample OTU richness; Colman et al. 2016). Similar to this study, a large proportion (up to 20%) of bacteria from a Chilean desert salt lake could not be identified to phylum level (Rasuk et al. 2016), indicating that in extreme environments, it is not unusual to find large proportions of unclassified bacteria in samples.
Conclusions
As acidic freshwater systems with high metal concentrations and relatively depauperate microbe communities, the Collie pit lakes could be considered extreme environments. However, as the macroinvertebrate data shows, the addition of even small amounts of carbon in the form of terrestrial nutrients altered biodiversity. In Lake WO5H, trees planted below the eventual water line added nutrients in the form of terrestrial leaf litter to the aquatic ecosystem. This can have profound implications for remediation, where connection to catchments can be a fundamental part of mine closure planning (Blanchette and Lund 2016). Drawing on the significant body of wetland restoration literature, sculpting the sides of pit lakes to form littoral zones for semi-aquatic plants can provide nutrients and habitat, but can also function as the ‘kidneys’ of lakes with poor water quality, filtering undesirable elements and stabilising banks (Mitsch 2012; Mitsch et al. 1998, 2012, 2015).
Macroinvertebrate and microbial assemblages differed across lakes and correlated with some similar water quality and biotic characteristics, particularly the metals in Lake WO5H. In this respect, the information provided by the macroinvertebrates and microbes was complementary. Ecological indicators are only useful if there are clear links between the taxon and ecological role (Kurtz et al. 2001). As many of the pit lake sediment microbes were unidentified, we could not even begin to speculate on ecological function, even of the most abundant taxon in the study. Nevertheless, technology is advancing quickly and microbes are attractive bio-indicators due to rapid sample collection and analysis. Future environmental impact programs are likely to include microbes as they become increasingly attractive assessment tools (Lau et al. 2015).
References
Anderson M, Gorley RN, Clarke RK (2008) Permanova+ for primer: guide to software and statistical methods. Primer-E Limited., Plymouth
ANZECC/ARMCANZ (2000) Australian and New Zealand guidelines for fresh and marine water quality. In: Australian and New Zealand Environment and Conservation Council and Agriculture and Resource Management Council of Australia and New Zealand, Canberra, pp 1–103
APHA (1998a) 4500-P J. Persulfate method for simultaneous determination of total nitrogen and total phosphorus. In: Standard methods for the examination of water and wastewater, American Public Health Assoc, American Water Works Assoc, and Water Environment Federation, Washington, DC
APHA (1998b) Standard methods for the examination of water and wastewater, 20th edn. APHA, Washington, DC
Australian Bureau of Meteorology (2019) Weather and climate data. http://www.bom.gov.au/climate/. Accessed 14 Jun
Bainton N, Holcombe S (2018) A critical review of the social aspects of mine closure. Resour Policy 59:468–478
Blanchette ML, Lund MA (2016) Pit lakes are a global legacy of mining: an integrated approach to achieving sustainable ecosystems and value for communities. Curr Opin Environ Sustain 23:28–34
Blanchette ML, Lund MA (2017) Biophysical closure criteria without reference sites: evaluating river diversions around mines. In: Proceedings, IMWA conference, vol I, pp 437–444. ISBN: 978-952-335-065-6
Blanchette ML, Pearson RG (2012) Macroinvertebrate assemblages in rivers of the Australian dry tropics are highly variable. Freshw Sci 31(3):865–881. https://doi.org/10.1899/11-068.1
Blanchette M, Pearson R (2013) Dynamics of habitats and macroinvertebrate assemblages in rivers of the Australian dry tropics. Freshw Biol 58:742–757
Blanchette ML, Lund M, Moore M, Short D (2019) Incorporating microbes into environmental monitoring and mine closure programs: river diversions as test beds. In: Proceedings of international mine water assoc (IMWA) conference, pp 645–652. ISBN: 978-5-91252-145-4
Campbell RS, Lind OT (1969) Water quality and aging of strip-mine lakes. J Water Pollut Control Fed 41(11):1943–1955
Cao Y, Larsen D, Thorne RS-J (2001) Rare species in multivariate analysis for bioassessment: some considerations. J N Am Benthol Soc 20(1):144–153
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6(8):1621
Carew ME, Pettigrove VJ, Metzeling L, Hoffmann AA (2013) Environmental monitoring using next generation sequencing: rapid identification of macroinvertebrate bioindicator species. Front Zool 10(1):1–15
Chou H, Pathmasiri W, Deese-spruill J, Sumner SJ, Jima DD, Funk DH, Jackson JK, Sweeney BW, Buchwalter DB (2018) The good, the bad, and the lethal: gene expression and metabolomics reveal physiological mechanisms underlying chronic thermal effects in mayfly larvae (Neocloeon triangulifer). Front Ecol Evol 6:27
Coe MW, Schmelz D (1972) A preliminary description of the physico-chemical characteristics and biota of three strip mine lakes, Spencer County, Indiana. Proc Indiana Acad Sci 82:184–188
Colman DR, Feyhl-Buska J, Robinson KJ, Fecteau KM, Xu H, Shock EL, Boyd ES (2016) Ecological differentiation in planktonic and sediment-associated chemotrophic microbial populations in Yellowstone hot springs. FEMS Microbiol Ecol. https://doi.org/10.1093/femsec/fiw168
Davis JA, Christidis F (1997) A guide to wetland invertebrates of Southwestern Australia. Western Australian Museum, Perth
Gammons CH, Harris LN, Castro JM, Cott PA, Hanna BW (2009) Creating lakes from open pit mines: processes and considerations, emphasis on northern environments. In: Canadian Technical Report of Fisheries and Aquatic Sciences, Ottawa, vol 2826
Gerbersdorf S, Wieprecht S (2015) Biostabilization of cohesive sediments: revisiting the role of abiotic conditions, physiology and diversity of microbes, polymeric secretion, and biofilm architecture. Geobiology 13(1):68–97
González-Toril E, Santofimia E, López-Pamo E, García-Moyano A, Aguilera Á, Amils R (2015) Comparative microbial ecology of the water column of an extreme acidic pit lake, Nuestra Señora del Carmen, and the Río Tinto basin (Iberian Pyrite Belt). Int Microbiol 17:225–233
Gooderham J, Tsyrlin E (2002) The Waterbug book: a guide to the freshwater macroinvertebrates of temperate Australia. CSIRO Publication, Clayton
Hawking JH, Theischinger G (1999) Dragonfly larvae (Odonata): a guide to the identification of larvae of Australian families and to the identification and ecology of final instar larvae from New South Wales. CRC for Freshwater Ecology, Canberra
Herlihy AT, Gerth WJ, Li J, Banks JL (2005) Macroinvertebrate community response to natural and forest harvest gradients in western Oregon headwater streams. Freshw Biol 50(5):905–919. https://doi.org/10.1111/j.1365-2427.2005.01363.x
Horton DJ, Theis KR, Uzarski DG, Learman DR (2019) Microbial community structure and microbial networks correspond to nutrient gradients within coastal wetlands of the Laurentian Great Lakes. FEMS Microbiol Ecol 95(4):1–17
Jones BF, Bowser CJ (1978) The mineralogy and related chemistry of lake sediments. Lakes. Springer, New York, pp 179–235
Kalin M, Wheeler WN (2013) Biological polishing of arsenic, nickel and zinc in an acidic lake and two alkaline pit lakes. In: Geller W, Schultze M, Kleinmann R, Wolkersdorfer C (eds) Acidic pit lakes: the legacy of coal and metal surface mines. Springer, Berlin, pp 387–408
Kampe H, Dziallas C, Grossart HP, Kamjunke N (2010) Similar bacterial community composition in acidic mining lakes with different pH and lake chemistry. Microb Ecol 60(3):618–627. https://doi.org/10.1007/s00248-010-9679-5
Karádi-Kovács K, Selmeczy GB, Padisák J, Schmera D (2015) Food, substrate or both? Decomposition of reed leaves (Phragmites australis) by aquatic macroinvertebrates in a large shallow lake (Lake Balaton, Hungary). Int J Limnol 51(1):79–88
Kean S (2009) Eco-alchemy in Alberta. Science 326(5956):1052–1055
Kurtz JC, Jackson LE, Fisher WS (2001) Strategies for evaluating indicators based on guidelines from the Environmental Protection Agency’s Office of Research and Development. Ecol Indic 1(1):49–60
Lau KE, Washington VJ, Fan V, Neale MW, Lear G, Curran J, Lewis GD (2015) A novel bacterial community index to assess stream ecological health. Freshw Biol 60(10):1988–2002
Leppänen JJ, Weckström J, Korhola A (2017) Multiple mining impacts induce widespread changes in ecosystem dynamics in a boreal lake. Sci Rep UK 7(1):10581
Lienhoop N, Messner F (2009) The economic value of allocating water to post-mining lakes in East Germany. Water Resour Manag 23(5):965–980
Lucheta A, Otero X, Macías F, Lambais M (2013) Bacterial and archaeal communities in the acid pit lake sediments of a chalcopyrite mine. Extremophiles 17(6):941–951
Lund M, Blanchette M (2018) Coal pit lake closure by river flow through: risks and opportunities. In: Report № 2018-3, Edith Cowan Univ, Mine water and environment research centre/centre for ecosystem management, Australian Coal Assoc Research Program
Lund MA, Blanchette ML, McCullough C (2014) Enhancing ecological values of coal pit lakes with simple nutrient additions and bankside vegetation. In: Report № C21038. Edith Cowan Univ mine water and environment research centre, centre for ecosystem management school of natural sciences, Australian Coal Assoc Research Program
Lund M, Blanchette M, Harkin C (2018) Seasonal river flow-through as a pit lake closure strategy: is it a sustainable option in a drying climate? In: Proceedings, 11th ICARD|IMWA|MWD conference: risk to opportunity, vol I, pp 34–41
Marchant R (2002) Do rare species have any place in multivariate analysis for bioassessment? J N Am Benthol Soc 21(2):311–313
McCullough CD, Lund MA (2006) Opportunities for sustainable mining pit lakes in Australia. Mine Water Environ 25(4):220–226
McCullough CD, Lund M, Zhao L (2009) Mine voids management strategy (I): pit lake resources of the Collie Basin. In: Department of Water Project Report MiWER/Centre for Ecosystem Management Report 14
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P (2012) An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6(3):610
Millar A, Mory A, Haig D, Backhouse J (2011) Collie coalfield and Lake Clifton—a field guide. Geological Survey of Western Australia, Record 2011/17, p 13
Miller GC, Lyons WB, Davis A (1996) Understanding the water quality of pit lakes. Environ Sci Technol 30(3):118A–123A. https://doi.org/10.1021/es9621354
Mitsch WJ (2012) What is ecological engineering? Ecol Eng 45:5–12
Mitsch WJ, Wu X, Nairn RW, Weihe PE, Wang N, Deal R, Boucher CE (1998) Creating and restoring wetlands. Bioscience 48(12):1019–1030
Mitsch WJ, Zhang L, Stefanik KC, Nahlik AM, Anderson CJ, Bernal B, Hernandez M, Song K (2012) Creating wetlands: primary succession, water quality changes, and self-design over 15 years. Bioscience 62(3):237–250
Mitsch WJ, Bernal B, Hernandez ME (2015) Ecosystem services of wetlands. Taylor & Francis, Routledge
Nagel R, Traub RJ, Allcock RJ, Kwan MM, Bielefeldt-Ohmann H (2016) Comparison of faecal microbiota in Blastocystis-positive and Blastocystis-negative irritable bowel syndrome patients. Microbiome 4(1):47
Naiman RJ (2013) Socio-ecological complexity and the restoration of river ecosystems. Inland Waters 3(4):391–410
Naiman RJ, Decamps H (1997) The ecology of interfaces: riparian zones. Annu Rev Ecol Syst 28(1):621–658
Rasuk MC, Fernández AB, Kurth D, Contreras M, Novoa F, Poiré D, Farías ME (2016) Bacterial diversity in microbial mats and sediments from the Atacama Desert. Microbiol Ecol 71(1):44–56
Rayment GE, Lyons DJ (2011) Soil chemical methods: Australasia, vol 3. CSIRO Publication, Clayton
Richardson J (1992) Food, microhabitat, or both? Macroinvertebrate use of leaf accumulations in a montane stream. Freshw Biol 27(2):169–176
Roch M, Nordin R, Austin A, McKean C, Deniseger J, Kathman R, McCarter J, Clark M (1985) The effects of heavy metal contamination on the aquatic biota of Buttle Lake and the Campbell River drainage (Canada). Arch Environ ConTox 14(3):347–362
Simonin M, Voss KA, Hassett BA, Rocca JD, Wang SY, Bier RL, Violin CR, Wright JP, Bernhardt ES (2019) In search of microbial indicator taxa: shifts in stream bacterial communities along an urbanization gradient. Environ Microbiol. https://doi.org/10.1111/1462-2920.14694
Smith M, Kay W, Edward D, Papas P, Richardson KSJ, Simpson J, Pinder A, Cale D, Horwitz P, Davis J (1999) AusRivAS: using macroinvertebrates to assess ecological condition of rivers in Western Australia. Freshw Biol 41(2):269–282
Stedman C (1988) 100 Years of Collie coal. Curtin University of Technology, Perth
Suter PJ (1999) Illustrated key to the Australian Caenid nymphs (Ephemeroptera: Caenidae). In: Presented at the taxonomic workshop at the Murray–Darling Freshwater Research Centre, Albury, 2–4 Feb 1999. Cooperative Research Centre for Freshwater Ecology
USEPA (1996) Method 3050B: acid digestion of sediments, sludges, and soils revision 2. Washington, DC
Vander Vorste R, Timpano AJ, Cappellin C, Badgley BD, Zipper CE, Schoenholtz SH (2019) Microbial and macroinvertebrate communities, but not leaf decomposition, change along a mining-induced salinity gradient. Freshw Biol 64(4):671–684
Watts CH (2002) Checklists and guides to the identification, to genus, of adult and larval Australian water beetles of the families Dytiscidae, Noteridae, Hygrobiidae, Haliplidae, Gyrinidae, Hydraenidae and the superfamily Hydrophiloidea (Insecta: Coleoptera). In: Presented at the taxonomy workshop, Lake Hume Resort, 5–6 Feb 2002, Cooperative Research Centre for Freshwater Ecology
Wielinga B (2009) Microbial Reactions. In: Castendyk D, Eary L (eds) Mine pit lakes: characteristics, predictive modelling, and sustainability, vol 3. Society for mining, metallurgy, and exploration. ACS Publications, Littleton, pp 147–157
Williams WD (1980) Australian freshwater life: the invertebrates of Australian inland waters. Macmillan Education AU, New York
Woodbury R (1998) Butte, Montana: the giant cup of poison. Time. http://content.time.com/time/magazine/article/0,9171,988063,00.html. Accessed 22 Oct 2019
Yergeau E, Michel C, Tremblay J, Niemi A, King TL, Wyglinski J, Lee K, Greer CW (2017) Metagenomic survey of the taxonomic and functional microbial communities of seawater and sea ice from the Canadian Arctic. Sci Rep 7:42242
Younger PL, Wolkersdorfer C (2004) Mining impacts on the fresh water environment: technical and managerial guidelines for catchment scale management. Mine Water Environ 23:s2–s80
Acknowledgements
The authors thank Digby Short, Colm Harkin, Michael Moore, Emily Evans (Premier Coal), and Paul Irving (Griffin Coal), as well as the ACARP board and administration for their support. ECU undergraduate students collected select data (biophysical characteristics, macroinvertebrates) and we thank the many ECU volunteers, students, and colleagues for their conversations and assistance in the field and laboratory. Finally, we wish to acknowledge the people of Collie, Western Australia for their trust and hospitality during the many years we have worked in their town.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Research was funded by the Australian Coal Association Research Program (ACARP) project C21038, Premier Coal and Griffin Coal. MLB is funded by ACARP and declares no conflict of interest.
Ethical approval
This research was conducted according to the Australian Code for the Responsible Conduct of Research and the Edith Cowan University Framework for the Responsible Conduct of Research.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10230_2019_647_MOESM1_ESM.docx
Benthic aquatic and riparian characteristics at Collie mine lake sites in April, 2013. Site abbreviations were ‘WO5H’ (Lake WO5H), ‘K’ (Lake Kepwari), and ‘S’ (Lake Stockton) followed by site number. All data are indices unless specified, and indices may not add to 100%. Methods from Blanchette and Pearson (2012, 2013), Blanchette and Lund (2017), Blanchette et al (2019). Benthic cover in macroinvertebrate collection transect (periphyton, aquatic plants, leaf litter, coarse particulate organic matter (CPOM)): 0; 0 % cover, 1; < 10 %, 2; 10-35 %, 3; 35-65 %, 4; 65-90 %, 5; > 90 %. Ground cover in 20 m riparian transect (grass, leaf litter, branches, exposed soil, trees, rushes, shrubs): 1; < 5 %, 2; 5-30 %, 3; 30-60 %, 4; 60-80 %, 5; 80-100 %. Bank slope index was 1; < 45°, 2; 45-70°, and 3; >70°. Bank mobility (20 m transect – undercutting, slumping and gullying) was 1; ‘none’, 2; ‘some’, and 3; ‘extensive.’ Sediment composition (based on a modified phi scale) was 1; sand-dominated (few rocks), 2; silt-dominated (few rocks), 3; sand and silt (few rocks), 4; complex rocky mix (no boulders), 5; boulders present. (DOCX 14 kb)
10230_2019_647_MOESM2_ESM.docx
Macroinvertebrate taxa collected from transects in Collie mine lakes, April, 2013. Data presented as range of individual number per transect with ‘-‘ indicating taxon not found in lake. Lake WO5H transect n = 13, Kepwari n = 21, Stockton n = 7. (DOCX 22 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Blanchette, M.L., Allcock, R., Gonzalez, J. et al. Macroinvertebrates and Microbes (Archaea, Bacteria) Offer Complementary Insights into Mine-Pit Lake Ecology. Mine Water Environ 39, 589–602 (2020). https://doi.org/10.1007/s10230-019-00647-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10230-019-00647-9