Abstract
Background
High density of tumor-infiltrating lymphocyte (TIL) is known to be associated with prolonged survival time, whereas tumoral-L1 hypomethylation has been associated with shortened survival time in patients with gastric cancer (GC). Since L1-methylation level is high in lymphocytes, higher density of TIL could lead to higher measurement of L1-methylation level in cancer tissues which contain cancer cells as well as non-neoplastic cells, including TIL. Putative interaction of TIL in the relationship between L1-methylation level and survival led us to explore combinatory statuses of tumoral-L1-methylation level and TIL density as a prognostic marker in GC.
Methods
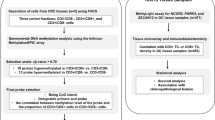
TIL and tumoral-L1-methylation level were measured in advanced GC samples (n = 491), using CD3 immunohistochemistry and pyrosequencing-methylation analysis, respectively. TIL density was measured in tumor center and invasive front areas.
Results
TIL density correlated with tumoral-L1-methylation level but the relationship was weak. Combinatory statuses of L1-methylation level and CD3 TIL density were found to be statistically significant in survival analysis. Multivariate analysis revealed that the relationship between combinatory statuses and survival was independent. Prognostic value of the combinatory statuses at invasive front was significant in an independent set.
Conclusions
Our findings indicate that tumoral-L1-methylation level is correlated with TIL density and that combinatory statuses might help to find a subset of GCs with worse clinical outcome in GCs with low-L1-methylation status or a subset of GCs with better clinical outcome in GCs with high-L1-methylation status.
Similar content being viewed by others
Introduction
5-Methylcytosine constitutes approximately 0.9% of human-genomic DNA and a decrease in the DNA-methylation content is a common finding in human cancers with exceptions such as renal-cell carcinoma and thyroid cancer [1]. Long interspersed nuclear element-1 (L1) comprises approximately 17% of human-genomic DNA and is repeated half a million times in the human genome in a dispersed manner. L1 has a high density of CpG dinucleotides in its 5′ untranslated region, and these CpG sites are usually hypermethylated in normal cells. However, 85% of L1 do not contain their 5′ sequences because early termination occurs, usually in the reverse transcription process, and the resultant reverse-transcribed DNA does not contain 5′ untranslated region. Nonetheless, methylation levels of 5′ untranslated region measured by the MethyLight assay or the pyrosequencing-methylation assay have been shown to correlate with the genomic DNA-methylation level measured by high performance liquid chromatography [2, 3]. Such a correlation provides basis for the utilization of L1-methylation level as surrogate marker for genomic DNA-methylation content. In many human-epithelial malignancies, tumoral-L1 hypomethylation has been demonstrated to be associated with a worse clinical outcome, including patients with gastric cancer (GC) or colorectal cancer [4,5,6,7,8]. Thus, tumoral-L1 hypomethylation is gaining attention as a prognostic biomarker for GC or colorectal cancer.
Tumor infiltrating lymphocytes (TILs) protect hosts from aggressive behavior of malignant cells, and GCs with a high density of TILs exhibit better clinical outcomes compared with GCs with a low density of TILs [9, 10]. Because the L1-methylation level is higher in leukocytes than in tumor tissues [11], tumor tissues with higher density of TILs are more likely to show higher measured values of L1-methylation levels when the tumor tissues, a mixture of tumor cells and non-neoplastic cells, are assayed for their L1-methylation levels. Considering that TILs and the methylation level of L1 are both prognostic factors in GC [4, 9], there is a concern that the relationship between tumoral-L1 hypomethylation and worse clinical outcome might be a spurious one that depends on the density of TILs in the cancer tissue. In other words, the possibilities that the worse clinical outcome observed in tumors with a low level of L1 methylation is related to the low density of TILs and that the better clinical outcome in tumors with a high level of L1 methylation might result from a high density of TILs need to be ruled out. However, no study has investigated this issue.
We hypothesized that the prognostic implication of the tumoral-L1-methylation level might be affected by TIL density. In the current study, we sought to determine whether tumoral-L1-methylation level correlated with the TIL density. Furthermore, we sought to elucidate whether the combination of the tumoral-L1-methylation level and the TIL density might be useful in identifying subsets of patients with GCs with worse or better survival. We analyzed 491 cases of advanced gastric cancer (AGC) for their level of L1 methylation and TIL density using pyrosequencing and tissue microarray (TMA)-based CD3 immunohistochemistry, respectively. The TIL density was correlated with the L1-methylation level and clinicopathological features, including prognosis. The combination of the L1-methylation level and the TIL density were examined for their relationship with survival. Survival analysis in an independent set of AGCs validated the prognostic value of the combinatory statuses.
Materials and methods
Patient selection and specimen
Formalin-fixed–paraffin-embedded (FFPE) archival-tissue samples of AGC (n = 491) were obtained from patients who had undergone surgical resection at Seoul National University Hospital, Seoul, Korea between 2007 January and 2008 December. Patients were included in the study if they met the following criteria: age at diagnosis > 18 years, AGC, adenocarcinoma histology, and availability of FFPE-cancer tissues. Patients with the following criteria were excluded from the study: history of neoadjuvant therapy for GC, concurrent malignancy of other organs (except for papillary thyroid cancer) within 5 years, or a refusal to participate in the molecular study. Clinical and histological information was obtained from the electronic medical records, including Lauren histology, differentiation grade, tumor location within the stomach, lymphatic embolus, venous invasion status, and perineural invasion status, tumor-node-metastasis (TNM) stage (according to American Joint Committee on Cancer, 7th edition). When more than one primary tumor was present in the resected stomach specimen, the data were retrieved from higher T-stage tumor or, if the multiple primary tumors were of the same tumor stage, the larger tumor. Lymphocytic reactions were evaluated with semiquantitative criteria. For the validation study, we retrieved another set of AGCs (n = 300) which were excised at Seoul National University Hospital during 2009. The selection of the validation set was performed under the same criteria for the testing set. This study, including waiver of informed consent, was approved by the Institutional Review Board (IRB No.: 1312-051-542).
Pyrosequencing methylation assay of L1
Through microscopic examination of tissue slides, up to 1 cm2 samples from tumor areas that were the most cellular and represented the most prevalent histology of the individual case were marked. Unstained-tissue slides were marked on the corresponding areas and then deparaffinized with xylene. The marked areas were scraped and collected into microtubes containing tissue lysis buffer and proteinase K. After being kept at 95 °C for 24 h, the microtubes were subjected to thermal degradation at 95 °C for 30 min to decrease interstrand DNA-crosslinking induced by formalin-fixation [12]. The lysed tissue solutions were centrifuged and the supernatants were transferred to new microtubes. Bisulfite-modification of DNA samples was performed using EZ-DNA methylation kit (Zymo Research, Orange, CA, USA). The modified-DNA samples were subjected to PCR amplification using the same primers designed by Dr. Issa’s group for the pyrosequencing-methylation assay of L1 [13]. A subsequent pyrosequencing assay was conducted as described previously [14]. The reproducibility of the bisulfite conversion in FFPE DNA, including the mean of bisulfite-to-bisulfite standard deviation and the mean of run-to-run standard deviation for the pyrosequencing, has been demonstrated in our previous study [14]. Using the median value of the L1-methylation level as cut-off value, the tumors were classified into low- and high-L1 methylation groups.
Tissue microarray and CD3 immunohistochemistry
After microscopic examination, three tissue cores (core diameter, 2 mm) with representative histology were selected from both the tumor center (one core per patient sample) and invasive-front regions (two cores from patient sample). For the invasive-front region, cores were located within the tumor area inside the tumor boundary. Two cores were obtained from the invasive-front region because the TIL density was more variable in the invasive-front region than in the tumor-center region. TMA was constructed with three tissue cores from each case. Four-micrometer-thick sections of TMA tissue blocks were stained with the anti-CD3 antibody (1:200, Dako, Glostrup, Denmark). CD3-positive cells were counted using the open source software QuPath [15]. The output was CD3 TIL density (the number of CD3-positive cells per mm2 of tissue) for each TMA core. For each case, the CD3 TIL density from one core at the tumor center and the mean value of the CD3 TIL densities from two cores at the invasive front were determined. With the cut-off values set at the median value of CD3 TIL density at the tumor center or invasive front, the tumors were classified into low-CD3 density at the tumor center (CD3-lowcent) and high-CD3 density at the tumor center (CD3-highcent) groups or into low-CD3 density at the invasive front (CD3-lowinv) and high-CD3 density at the invasive front (CD3-highinv) groups, respectively.
Epstein–Barr virus in situ hybridization and microsatellite instability test
Epstein–Barr virus (EBV)-encoded RNA in situ hybridization was performed on sections of the TMA, as described previously, to detect the presence of EBV in the cancer cells [16]. Using capillary electrophoresis, the instability at five microsatellite loci (BAT25, BAT26, D2S123, D5346, and D17S250) was examined to determine the microsatellite instability (MSI) status. Tumors with a high level of MSI (MSI-high or MSI-H) were defined as tumors showing instability at two or more loci, whereas tumors were defined as microsatellite-stable (MSS) tumors when they exhibited instability at one or none of the five loci.
Statistical analysis
The statistical analysis was performed with SPSS software for Windows, version 25.0 (IBM, Chicago, IL, USA). Using the Shapiro–Wilk test, the normality of the distributions of the CD3 densities and L1-methylation levels were determined. The P values were less than 0.001, which rejected the null hypothesis that the data was normally distributed. In a previous study, we found that L1-methylation levels were not normally distributed [14]. Because of these findings, both parametric tests and non-parametric tests were performed to identify any difference in the means between two or more groups. Student’s t test and the Mann–Whitney test were used for the comparison of means between two groups and ANOVA test and the Kruskal–Wallis test were used for the comparison of means among three or more groups. Both the Pearson correlation and Spearman tests were used to determine the correlations between variables. Overall survival (OS) was defined as the time from the date of surgery to the date of death. Cancer-specific survival (CSS) was defined as the time from the date of surgery to the date of death caused by AGC. A Kaplan–Meier curve with a log-rank test was performed for visualization of categorized variables. A Cox proportional hazard model was used to calculate the hazard ratio (HR), and baseline characteristics were adjusted for covariates (age, tumor subsite, T stage, N stage, M stage, lymphatic emboli, venous invasion, perineural invasion, Lauren classification, lymphocytic reaction, and adjuvant therapy) that were found to be significant in the univariate survival analysis (P < 0.05). Subsequently, a backward stepwise elimination was carried out with P = 0.05 as a threshold to select variables for the final model.
Results
Demographical features and their relationships with CD3 TIL density
The demographical findings are summarized in Table 1. The CD3 TIL density was higher in the center region than in the invasive-front region (median, 710 vs. 203; P value, < 0.001). A lower density of CD3 TIL in the tumor-center region was associated with intestinal type histology, higher T, N, or M category, higher TNM stage, lymphatic emboli, venous invasion, EBV-negative/MSS molecular subtype, no lymphocytic reaction, and application of adjuvant chemotherapy (Table 1).
Correlations between CD3 density and L1-methylation level
Neither the CD3 densities in the tumor center and invasive front regions nor L1-methylation levels were normally distributed. To identify whether there was a positive correlation between CD3 density and L1-methylation level, both Pearson correlation and Spearman test were performed. Correlation coefficients for Pearson correlation and Spearman test were 0.253 and 0.283 for the tumor-center region and 0.154 and 0.099 for the invasive-front region. Although the association between CD3 density and L1-methylation level were statistically significant, the strength of the relationships was weak (Fig. S1). Because the CD3 density was different between GCs of different histologic types, subgroup analysis according to histologic types was performed to determine the correlation between CD3 density and L1-methylation levels. For GCs of the intestinal type, the correlation coefficients for Pearson test and Spearman test were 0.331 and 0.336, respectively, between the CD3 density of the center region and the L1-methylation level. However, the correlation coefficients were 0.169 and 0.197, respectively, for GCs of the diffuse type and 0.239 and 0.223, respectively, for GCs of the mixed type (Table S1).
Prognosis of patients with gastric cancer according to the combination of L1-methylation level and CD3 density
Although the CD3 TIL density differed between the tumor center and the invasive-front regions, a low density of CD3 TIL was associated with a worse clinical outcome regardless of whether the density was measured in the tumor center or the invasive front region (Fig. 1 and Fig. S2 for OS and CSS, respectively). Because a low-L1 methylation level was also significantly associated with worse clinical outcomes (Fig. S3), the combination of the L1 methylation and the CD3 TIL density was able to identify subgroups of patients with different clinical outcomes among patients with GC with a low-L1 methylation level. The combination of the L1-methylation level and the TIL density at the tumor center was found to be significant in the Kaplan–Meier curves and log-rank test for OS and CSS (Fig. 2a, Fig. S4a). Additionally, statistical significance was observed in the Kaplan–Meier survival analysis for the combination of the L1-methylation level and the TIL density at the invasive-front region (Fig. 2b, Fig. S4b). Multivariate analysis was performed with inclusion of prognostic parameters that were found to be significantly associated with survival in univariate analysis, including age at surgery, tumor subsite, T category, N category, M category, perineural invasion, lymphatic invasion, venous invasion, Lauren classification, lymphocytic reaction, and adjuvant chemotherapy. Multivariate analyses revealed that both the combination of the L1-methylation level and the CD3 TIL density at the tumor center and the combination of the L1-methylation level and the CD3 TIL density at the invasive front were significant factors for OS and CSS (Table 2 and Table S2, respectively). To validate prognostic significance of both combinations, survival analyses were performed in an independent set of advanced gastric cancers. Both combinatory statuses were statistically significant (Fig. 3, Fig. S5), but multivariate analyses in an independent set of AGCs revealed that the combination of the L1-methylation level and the CD3 density at the invasive front was a significant prognostic parameter and that combinatory statuses of L1 methylation and CD3 density at the tumor center was not a significant prognostic parameter for OS (Table 3 and Table S3).
Kaplan–Meier survival curves for four subgroups generated by a combination of L1-methylation levels and CD3 TIL densities of overall survival. a Survival curves for the combination of the L1-methylation level and the CD3 TIL density at the tumor center. b Survival curves for the combination of the L1-methylation level and the CD3 TIL density at the invasive front
Discussion
In the literature, there is an accumulating series of studies indicating that tumoral-L1 hypomethylation is associated with poor clinical outcome in patients with epithelial malignancy, including GC [4, 6, 17], esophageal-squamous cell carcinoma [18, 19], colorectal cancer [5], hepatocellular carcinoma [20], cholangiocarcinoma [21], and non-small cell lung cancer [22]. However, contrasting results are also reported in pancreatic adenocarcinoma and pulmonary adenocarcinoma. In pancreatic adenocarcinoma [23], no prognostic difference depending on tumoral-L1-methylation status has been reported, while a better prognosis in association with tumoral-L1 hypomethylation has been demonstrated in stage Ia pulmonary adenocarcinoma [24]. Such a discrepancy might be related to the difference in tissue types, histologic subtype, or molecular subtype. Molecular subtype is associated with variation in the L1- methylation level across tumors. In colorectal cancer, the tumoral-L1 methylation level is higher in MSI-H tumors than in MSS tumors [11, 25], while in GCs, the tumoral-L1 methylation level is higher in EBV-positive tumors than in MSI-H tumors and in MSI-H tumors than in MSS tumors [17]. Another confounding factor in the correlation between tumoral-L1 hypomethylation and patient survival is tumor location within the large bowel; the association of tumoral-L1 hypomethylation with cancer-specific survival was significant in the proximal colon but not in the distal colon and in the rectum [26]. The hazard ratio of tumoral-L1 hypomethylation for cancer-specific death was found to decrease from the right colon to the left colon to the rectum [26]. A potential confounding factor in the relationship between the tumoral-L1-methylation level and patient survival is TIL density. Because L1 methylation is higher in TILs than in cancer cells [11], the L1methylation level is expected to be higher in tumors with a high-TIL density than in tumors with a low-TIL density. Thus, it has been unclear whether the better survival in association with a high level of L1 methylation is due to the high-TIL density. In the present study, we investigated the effects of the interaction between the L1-methylation level and the TIL density on OS and CSS. We found that the combination of the L1-methylation level and the CD3 TIL density, regardless of whether the latter was in the tumor center and the invasive front, were both significant prognostic parameters for GC in univariate analysis of testing and validation sets for OS and CSS (Table S3). However, multivariate analysis revealed that prognostic significance of the combination of the L1-methylation level and the TIL density at the invasive front was reproducible in both testing- and validation-sets. Therefore, the combination of the L1-methylation level and the TIL density at the invasive front enabled further identification of subgroups with better prognosis in patients with GC with a low-L1 methylation status or worse prognosis in patients with GC with a high-TIL density.
Recently, a study showed that TIL density was inversely correlated with L1-RNA expression and the number of L1 retro-transposition events in gastrointestinal tract cancers [27]. Although the study did not analyze methylation levels of L1, increased expression of L1 RNA might represent a decreased level of L1 methylation in the genomic sequences and thus tumor tissues with a higher TIL density might harbor high level of L1 methylation in tumor cells. In other words, the measured high level of L1 methylation in tumor tissues with high TIL density might not be entirely due to the high density of TIL. Three possibilities can be considered for the reason why tumor cells surrounded by a high-TIL density maintain a high level of L1 methylation. First, a high density of TILs might provide methylation cues that lead to elevated-DNA methyltransferase levels and subsequently enhanced-methylation levels of the genomic DNAs in tumor cells. Second, tumor cells maintain a high level of DNA methylation due to their own inherent molecular features which are also linked with recruitment of TIL, e.g., EBV-positive gastric cancer and CIMP-induced MSI-high GC. These two molecular subtypes show high levels of L1 methylation regardless of TIL density but usually also exhibit high-TIL density. Third, a high density of TILs has a tendency to be associated with a high density of intratumoral-macrophage infiltration [28], and such a high density of chronic inflammatory cell infiltration has been reported to be linked with increased methylation in non-neoplastic gastric epithelial cells [29]. A recent study has shown that the increased infiltration of tumor-associated macrophages leads to the elevation of the level of DNA methyltransferases and enhanced-DNA methylation in gastric cancer cells [30].
Because tumor tissue is a mixture of tumor cells and non-neoplastic cells, including immune cells, inflammatory cells, and fibroblasts, the proportion of non-neoplastic cells in the dissected tumor areas ranged from 10 to 90% with a median of 50% in the present study. In a previous study, we found that a low L1-methylation status was prognostic in the subset of patients with tumor purity \(\ge\)50% but the association between the L1-methylation status and survival in the subset of patients with tumor purity < 50% was marginally significant [14]. However, in the present study, the combination of the L1 methylation and the TIL density statuses had persistent prognostic power regardless whether tumor purity was \(\ge\)50% or < 50% (Fig. S6). In particular, the survival curve of the patients in the L1-low/CD3-lowcent subgroup was distinct from those in the other three subgroups and demonstrated the worst clinical behavior regardless of tumor purity. In contrast, the combination of the L1 methylation and the TIL density at the invasive front area displayed contrasting survival curves between those with tumor purity \(\ge\) 50% and < 50%. For the subset of tumor purity \(\ge\) 50%, the L1-low/CD3-lowinv subgroup had the worst survival, and the other three subgroups exhibited similar survival curves; however, for the subset of tumor purity values < 50%, the L1-high/CD3-highinv had the best survival curve, and the other three subgroups showed similar survival curves.
In the present study, we found that survival curves for the combination of the L1-methylation level and the CD3 TIL density were different depending on whether L1-methylation status was combined with TIL density at the tumor center or that at the invasive front. The combination of the L1-methylation level and the CD3 TIL density at the tumor center tended to yield one worse subgroup (L1-low/CD3-lowcent) and three similarly better subgroups on the aspect of survival, whereas the combination of the L1-methylation level and the CD3 TIL density at the invasive front tended to yield one worst subgroup (L1-low/CD3-lowinv), two intermediate subgroups (L1-low/CD3-highinv and L1-high/CD3-lowinv), and one best subgroup (L1-high/CD3-highinv). The pattern of one worse and three better survival curves for the statuses of L1 methylation and TIL density at tumor center was persistent regardless of tumor purity \(\ge\) 50% or < 50%, whereas the pattern of one worst survival curve, two intermediate survival curves, and one best survival curve for the combination of L1 methylation and TIL density at invasive front was not observed when survival analysis was performed to AGCs with tumor purity \(\ge\) 50% only or < 50% only. As mentioned above, survival analysis for the combination of L1 methylation and TIL density at the invasive front disclosed one worse survival curve and three better survival curves in AGCs with tumor purity ≥ 50% and one better survival curve and three worse survival curves in AGCs with tumor purity < 50%. Survival curves for the combination of the L1 methylation and CD3 TIL density at the invasive center differed depending on tumor purity. Thus, our findings suggest that prognostic impact of the combination of the L1-methylation level and TIL density at invasive center on patients with GC might be influenced by tumor purity.
To explore whether the prognostic value of the L1 methylation on OS is influenced by TIL status, we tested the statistical significance of interaction term of L1 methylation and CD3 TIL density. For both tumor center and invasion front, the cross product of L1 methylation and CD3 TIL density was not significantly associated with OS (P = 0.224 and P = 0.282, respectively) while L1 methylation remained as an independent prognostic factor (P < 0.05). Next, we evaluated the variance inflation factor (VIF) between L1 methylation and CD3 TIL density, and the VIF values were 1.03 and 1.00 for tumor center and invasion front, respectively. These results indicate that the L1 methylation and CD3 TIL density did not show significant co-linearity on OS in our dataset.
In conclusion, we found that the tumoral-L1-methylation level was correlated with TIL density but the association was weak. The combination of tumoral-L1-methylation level and TIL density at the invasive front were found to contribute to identification of subsets with better or worse prognosis in GCs with low-L1-methylation status or low-TIL density.
References
Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, Thong-ngam D, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23(54):8841–6. https://doi.org/10.1038/sj.onc.1208137.
Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33(21):6823–36. https://doi.org/10.1093/nar/gki987.
Lisanti S, Omar WA, Tomaszewski B, De Prins S, Jacobs G, Koppen G, et al. Comparison of methods for quantification of global DNA methylation in human cells and tissues. PLoS O ne. 2013;8(11):e79044. https://doi.org/10.1371/journal.pone.0079044.
Shigaki H, Baba Y, Watanabe M, Murata A, Iwagami S, Miyake K, et al. LINE-1 hypomethylation in gastric cancer, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastric Cancer. 2013;16(4):480–7. https://doi.org/10.1007/s10120-012-0209-7.
Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Chan AT, Schernhammer ES, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100(23):1734–8. https://doi.org/10.1093/jnci/djn359.
Bae JM, Shin SH, Kwon HJ, Park SY, Kook MC, Kim YW, et al. ALU and LINE-1 hypomethylations in multistep gastric carcinogenesis and their prognostic implications. Int J Cancer. 2012;131(6):1323–31. https://doi.org/10.1002/ijc.27369.
Iwagami S, Baba Y, Watanabe M, Shigaki H, Miyake K, Ishimoto T, et al. LINE-1 hypomethylation is associated with a poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Ann Surg. 2013;257(3):449–55. https://doi.org/10.1097/SLA.0b013e31826d8602.
Kim Y, Wen X, Jeong S, Cho NY, Kim WH, Kang GH. Combinatory low methylation statuses of SAT-alpha and L1 are associated with shortened survival time in patients with advanced gastric cancer. Gastric Cancer. 2018;22(1):37–47. https://doi.org/10.1007/s10120-018-0852-8.
Lee HE, Chae SW, Lee YJ, Kim MA, Lee HS, Lee BL, et al. Prognostic implications of type and density of tumour-infiltrating lymphocytes in gastric cancer. Br J Cancer. 2008;99(10):1704–11. https://doi.org/10.1038/sj.bjc.6604738.
Zheng X, Song X, Shao Y, Xu B, Chen L, Zhou Q, et al. Prognostic role of tumor-infiltrating lymphocytes in gastric cancer: a meta-analysis. Oncotarget. 2017;8(34):57386–98. https://doi.org/10.18632/oncotarget.18065.
Estecio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS O ne. 2007;2(5):e399. https://doi.org/10.1371/journal.pone.0000399.
Wen X, Jeong S, Kim Y, Bae JM, Cho NY, Kim JH, et al. Improved results of LINE-1 methylation analysis in formalin-fixed, paraffin-embedded tissues with the application of a heating step during the DNA extraction process. Clin Epigenetics. 2017;9:1. https://doi.org/10.1186/s13148-016-0308-0.
Tellez CS, Shen L, Estecio MR, Jelinek J, Gershenwald JE, Issa JP. CpG island methylation profiling in human melanoma cell lines. Melanoma Res. 2009;19(3):146–55.
Kim Y, Wen X, Jeong S, Cho NY, Kim WH, Kang GH. Combinatory low methylation statuses of SAT-alpha and L1 are associated with shortened survival time in patients with advanced gastric cancer. Gastric Cancer. 2019;22(1):37–47. https://doi.org/10.1007/s10120-018-0852-8.
Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878. https://doi.org/10.1038/s41598-017-17204-5.
Kang GH, Lee S, Cho NY, Gandamihardja T, Long TI, Weisenberger DJ, et al. DNA methylation profiles of gastric carcinoma characterized by quantitative DNA methylation analysis. Lab Investig. 2008;88(2):161–70. https://doi.org/10.1038/labinvest.3700707.
Song YS, Kim Y, Cho NY, Yang HK, Kim WH, Kang GH. Methylation status of long interspersed element-1 in advanced gastric cancer and its prognostic implication. Gastric Cancer. 2016;19(1):98–106. https://doi.org/10.1007/s10120-015-0463-6.
Furlan C, Polesel J, Barzan L, Franchin G, Sulfaro S, Romeo S, et al. Prognostic significance of LINE-1 hypomethylation in oropharyngeal squamous cell carcinoma. Clin Epigenetics. 2017;9:58. https://doi.org/10.1186/s13148-017-0357-z.
Baba Y, Watanabe M, Murata A, Shigaki H, Miyake K, Ishimoto T, et al. LINE-1 hypomethylation, DNA copy number alterations, and CDK6 amplification in esophageal squamous cell carcinoma. Clin Cancer Res. 2014;20(5):1114–24. https://doi.org/10.1158/1078-0432.CCR-13-1645.
Harada K, Baba Y, Ishimoto T, Chikamoto A, Kosumi K, Hayashi H, et al. LINE-1 methylation level and patient prognosis in a database of 208 hepatocellular carcinomas. Ann Surg Oncol. 2015;22(4):1280–7. https://doi.org/10.1245/s10434-014-4134-3.
Jeong S, Lee K, Wen X, Kim Y, Cho NY, Jang JJ, et al. Tumoral LINE-1 hypomethylation is associated with poor survival of patients with intrahepatic cholangiocarcinoma. BMC Cancer. 2017;17(1):588. https://doi.org/10.1186/s12885-017-3595-8.
Imperatori A, Sahnane N, Rotolo N, Franzi F, Nardecchia E, Libera L, et al. LINE-1 hypomethylation is associated to specific clinico-pathological features in stage I non-small cell lung cancer. Lung Cancer. 2017;108:83–9. https://doi.org/10.1016/j.lungcan.2017.03.003.
Yamamura K, Kosumi K, Baba Y, Harada K, Gao F, Zhang X, et al. LINE-1 methylation level and prognosis in pancreas cancer: pyrosequencing technology and literature review. Surg Today. 2017;47(12):1450–9. https://doi.org/10.1007/s00595-017-1539-1.
Rhee YY, Lee TH, Song YS, Wen X, Kim H, Jheon S, et al. Prognostic significance of promoter CpG island hypermethylation and repetitive DNA hypomethylation in stage I lung adenocarcinoma. Virchows Arch. 2015. https://doi.org/10.1007/s00428-015-1749-0.
Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122(12):2767–73. https://doi.org/10.1002/ijc.23470.
Mima K, Nowak JA, Qian ZR, Cao Y, Song M, Masugi Y, et al. Tumor LINE-1 methylation level and colorectal cancer location in relation to patient survival. Oncotarget. 2016;7(34):55098–109. https://doi.org/10.18632/oncotarget.10398.
Jung H, Choi JK, Lee EA. Immune signatures correlate with L1 retrotransposition in gastrointestinal cancers. Genome Res. 2018. https://doi.org/10.1101/gr.231837.117.
Kim KJ, Wen XY, Yang HK, Kim WH, Kang GH. Prognostic implication of M2 macrophages are determined by the proportional balance of tumor associated macrophages and tumor infiltrating lymphocytes in microsatellite-unstable gastric carcinoma. PLoS O ne. 2015;10(12):e0144192. https://doi.org/10.1371/journal.pone.0144192.
Hur K, Niwa T, Toyoda T, Tsukamoto T, Tatematsu M, Yang HK, et al. Insufficient role of cell proliferation in aberrant DNA methylation induction and involvement of specific types of inflammation. Carcinogenesis. 2011;32(1):35–41. https://doi.org/10.1093/carcin/bgq219.
Wang HC, Chen CW, Yang CL, Tsai IM, Hou YC, Chen CJ, et al. Tumor-associated macrophages promote epigenetic silencing of gelsolin through DNA methyltransferase 1 in gastric cancer cells. Cancer Immunol Res. 2017;5(10):885–97. https://doi.org/10.1158/2326-6066.
Funding
This work was funded by a Grant from the National Research Foundation (NRF) funded by the Korean Ministry of Science, ICT and Future Planning (2016M3A9B6026921), a Grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute funded by the Korean Ministry of Health and Welfare (HI14C1277), and a Grant from SNUH Research Fund [0420180210 (2018-1212)], and Seegene Medical Foundation Scholarship fund (Grant no. 2019-001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10120_2019_1025_MOESM1_ESM.tif
Fig. S1. Scatter plots correlating the L1 methylation level and the CD3 TIL density at the tumor center (a, c, e, & g) or the CD3 TIL density at the invasive front (b, d, f, & h). (a) & (b) overall gastric cancer cases, (c) & (d) gastric cancer of the intestinal type, (e) & (f) gastric cancers of the diffuse type, and (g) & (h) gastric cancers of the mixed type (TIFF 1611 kb)
10120_2019_1025_MOESM2_ESM.tif
Fig. S2. Kaplan-Meier survival curves for low and high CD3 TIL density subgroups of cancer-specific survival. (a) Survival curves for CD3 TIL density at the tumor center. (b) Survival curves for CD3 TIL density at the invasive front (TIFF 577 kb)
10120_2019_1025_MOESM3_ESM.tif
Fig. S3. Kaplan-Meier survival curves for L1 methylation status of (a) overall survival and (b) cancer-specific survival (TIFF 481 kb)
10120_2019_1025_MOESM4_ESM.tif
Fig. S4. Kaplan-Meier survival curves for four subgroups generated by a combination of L1 methylation levels and CD3 TIL densities of cancer-specific survival. (a) Survival curves for the combination of the L1 methylation level and the CD3 TIL density at the tumor center. (b) Survival curves for the combination of the L1 methylation level and the CD3 TIL density at the invasive front (TIFF 701 kb)
10120_2019_1025_MOESM5_ESM.tif
Fig. S5. Kaplan-Meier survival curves for four subgroups generated by a combination of L1 methylation levels and CD3 TIL densities of cancer-specific survival in the validation set. (a) Survival curves for the combination of the L1 methylation level and the CD3 TIL density at the tumor center. (b) Survival curves for the combination of the L1 methylation level and the CD3 TIL density at the invasive front (TIFF 744 kb)
10120_2019_1025_MOESM6_ESM.tif
Fig. S6. Kaplan-Meier survival curves for the combination of the L1 methylation level and the CD3 TIL density at the tumor center according to tumor purity. (a) Survival curve for the combination of the L1 methylation level and the CD3 TIL density at the tumor center in gastric cancers with ≥50% tumor purity. (b) Survival curve for the combination of the L1 methylation level and the CD3 TIL density at the invasive front in gastric cancers with ≥50% tumor purity. (c) Survival curve for the combination of the L1 methylation level and the CD3 TIL density at the tumor center in gastric cancers with <50% tumor purity. (d) Survival curve for the combination of the L1 methylation level and the CD3 TIL density at the invasive front in gastric cancers with <50% tumor purity (TIFF 424 kb)
Rights and permissions
About this article
Cite this article
Kim, Y., Rhee, YY., Wen, X. et al. Combination of L1 methylation and tumor-infiltrating lymphocytes as prognostic marker in advanced gastric cancer. Gastric Cancer 23, 464–472 (2020). https://doi.org/10.1007/s10120-019-01025-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-019-01025-8