Abstract
The light-gated dimeric cation channel channelrhodopsin-2 (ChR2) has been established as one of the most important optogenetic tools. During its functional cycle, ChR2 undergoes conformational changes, the most prominent ones include a movement of transmembrane helix B. In the present work, we assign this movement to a trapped photocycle intermediate using DEER spectroscopy combined with sample illumination inside the microwave resonator, allowing trapping and relaxation of defined ChR2 intermediates at different temperatures between 180 and 278 K. Intradimer distances measured between spin-labeled positions 79 located in helix B of ChR2 in the dark state and upon light activation and relaxation at 180 K were similar. In contrast, light activation at 180 K and 30 min relaxation at between 230 and 255 K results in significant changes of the distance distribution. We show that the light-induced movement of helix B is correlated with the presence of the P480 state of ChR2. We hypothesize that conformational changes occurring in this area are key elements responsible for desensitizing the channel for cation conduction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
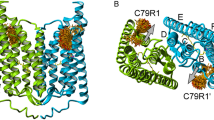
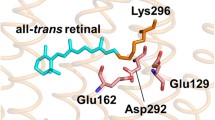
Channelrhodopsin-2 (ChR2) from the green alga Chlamydomonas reinhardtii is a light-gated cation channel, which depolarizes the cell interior upon light activation. It has been established as one of the most important optogenetic tools [1]. The primary sequence of ChR2 consists of 737 amino acids, whereof residues 316 to 737 build a cytosolic domain, which does not play a crucial role for the channel function [2]. The transmembrane part (amino acids 1 to 315) classifies ChR2 to the family of rhodopsins [3, 4]. This transmembrane part consists of seven almost parallel helices, labeled A to G (Fig. 1). ChR2 forms a dimer with dimerization sites on helices C and D [5, 6]. The X-ray structure of the closed ChR2 state [7] shows an electronegative cavity, surrounded by helices A, B, C, and G, as a part of the cation-conducting pathway, which is opened towards the extracellular side but occluded from the cytoplasmic side [6]. Residues within helices B and G seem to regulate the cation selectivity [8]. An inter-helical hydrogen-bond network forms two restriction areas (S63, Y70 (helix A), E90 (helix B), and N258 (helix G)) [6], the hydrogen-bond between C128 and D156 acts as a putative gate (DC gate) [9, 10].
Structural model of the ChR2 dimer in ribbon diagram representation (PDB: 6EID) the view parallel (A) and perpendicular (B) to the membrane plane shows the arrangement of the two protomers (green, blue). The two corresponding retinal chromophores are depicted in yellow. In (B) the seven transmembrane helices are labeled in one of the monomers from A to G. Spin label rotamers (orange, nitroxide oxygen atoms in red, stick representation) were calculated for positions 79 using MtsslWizard [11] and show the possible orientations of the spin label side chains. The observed movement of helix B (see text) is indicated by the gray arrows (color figure online)
The light-induced channel function of ChR2 is linked to its so-called photocycle. Activation of the channel by blue light leads to an isomerization around the C13–C14 bond of its chromophore retinal, which is covalently bound to K257 forming a protonated Schiff base [12,13,14,15].
The resulting red-shifted intermediate PK500 [16] (the nomenclature of the states was chosen as in [17]) decays to the blue-shifted intermediate PM390 in the µs time range. During this transition the retinal Schiff base becomes deprotonated. Reprotonation of the Schiff base occurs during the transition to the intermediate PN520 and leads to the conductive state of the protein [13]. Cation permeation starts to become inhibited by the recovered H-bonding network of the DC gate and protonation states of E90 and D156 so that the channel closes after about 10 ms [16]. More detailed investigations of the photocycle led to different extended photocycle models (e.g., [17,18,19,20,21]) with parallel branches including an intermediate P480. According to [17], P480 describes a light-adapted state of ChR2, which represents the photosensitive closed state of a second photocycle (syn-cycle). It is accumulated during continuous illumination [17] and can be stabilized by freeze trapping at 245 K [21].
Channel opening and closing require conformational changes. Structural alterations are well known for other rhodopsins, like bacteriorhodopsin [22, 23], sensory rhodopsin II [24, 25], and halorhodopsin [26]. IR spectroscopy disclosed that the pre-opening process, which is initiated by chromophore isomerization, is connected to an outward movement of the key residue E90 and is followed by a second step of inner gate opening [27]. DEER measurements using EPR-sensitive mutants recorded a movement of helix B [28, 29] accompanied by smaller structural changes of helix F in case of the decelerated C128T mutant [29]. Cw EPR measurements of a natural anion ChR labeled at the A–B loop supported the helix B movement upon light activation [30]. Cryo-electron microscopy measurements of two-dimensional crystals of the ChR2–C128T mutant confirmed a movement of helix F and a loss of order of helix B and indicated additional structural changes of helix G [31]. These helical movements were suggested to be correlated with the opening process of the protein but a rearrangement of helix B does not seem to be essential for the process of channel closure [28]. A connection between helical movements and the early stage of channel opening was also drawn by MD simulations [32], as these helical movements among others seem to be jointly responsible for water influx and cation permeation. Recent time-resolved serial femtosecond crystallography (TR-SFX) experiments on a chimeric ChR construct (C1C2) revealed an outward shift of helix C and the downward shift of helix G occurring early in the photocycle process [33]. The authors proposed that the observed conformational changes of these helices are the triggers that induce channel opening by eliciting conformational changes in the surrounding helices, such as helix B and helix F as observed in the ‘open’ conformation of the ChR2–C128T mutant [31].
The question still remains how the reported movements of helix B are correlated with the photocycle intermediates and channel function. Here, we present a combination of sample illumination inside the EPR resonator followed by thermal relaxation and DEER spectroscopy to study the intradimer distance between positions 79 in helix B of the light-activated ChR2 dimer. We show that the most prominent light-induced distance changes were observed in the presence of the trapped intermediate P480.
2 Materials and Methods
2.1 Expression and Purification
The ChR2 variant ChR2–C79R1was prepared according to published protocols [9, 28]. In brief, the expression of Chop2 (coding for amino acids residues 1 to 307 of GenBank accession number AF461397) in Pichia pastoris was induced in the presence of all-trans retinal by methanol added three times over a period of 30 h. The cells were harvested by centrifugation and resuspended to 30% wet weight in 20 mM sodium phosphate (pH 7.4), 100 mM NaCl, 2 mM EDTA, 1 mM PMSF and 5% (v/v) glycerol. Cell lysis was induced mechanically by glass beads (0.5 mm) in a bead-beater (BioSpec Products). The suspension was centrifuged for 15 min at 5000 g. The supernatant was centrifuged for 1 h at 100,000 g. The membranes were collected and homogenized in 20 mM HEPES/NaOH (pH 7.4), 100 mM NaCl and 0.2% (w/v) DM.
2.2 Site-Directed Spin Labeling
For site-directed spin labeling, the protein was bound to a Ni–NTA column and the cysteines were reduced with 5 mM DTT. After removal of DTT, the protein samples were incubated with 1 mM spin label [(1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate; MTSSL] for 12 to 20 h at 4 °C in darkness. Unbound spin label was removed by buffer exchange and the proteins were eluted with 1 M imidazole, before they were purified on a Superdex 200 column. Samples were concentrated by centrifugal filtration (Ultra-15, 10 kDa, Amicon).
2.3 Determination of Protein Concentrations and Labeling Efficiencies
The protein concentrations were determined from the optical absorbances measured at 280 nm and 460 nm using the extinction coefficients ε280 = 77,350 M−1 cm−1 [34] or ε460 = 45,000 M−1 cm−1 [7]. The extinction at 280 nm was used to calculate the concentration of the whole protein, whereas the extinction at 460 nm provided the information about the retinal bound protein concentration. The purity factor of ChR2 was determined from the ratio E280/E460, which theoretically reaches 1.7 for pure ChR2. Realistic experimental values are ~ 2.1 assuming 20% of bleached protein [34]. The purity factor of the prepared samples was calculated to be 2.8 indicating that ~ 75% of the protomers bound a retinal and thus denoting measurable light activation. The protein concentration of the samples used for DEER measurements were ~ 180 µM.
The spin labeling efficiencies were determined from double integration of the EPR spectra and comparison with the double integral of the EPR spectrum of spin label solution with known concentration. Spin labeling efficiencies of ChR2–C79R1 amounted to ~ 90%.
2.4 DEER Spectroscopy
Pulsed EPR experiments were performed at Q-band frequencies with a Bruker ELEXSYS 580 spectrometer equipped with a 150 W TWT amplifier (ASE, Fort Worth, Texas), a Bruker ER5106QT-2 resonator, and a continuous flow helium cryostat CF935 controlled by an Oxford Intelligent Temperature Controller ITC 503S. The samples were mixed with 20% D-glycerol as cryoprotector, 10 µL were filled into EPR tubes with 3 mm inner diameter and frozen immediately in liquid nitrogen. Measurements were performed using the four pulse DEER sequence and a two-step phase cycling. The resonator was critical coupled to Q ~ 400, the observer frequency was set to the center of the resonator dip and the pump frequency was shifted up by 50 MHz. The latter coincided with the absolute maximum of the nitroxide EPR spectrum, whereas the observer frequency coincided with the local maximum. All measurements were performed at 50 K with a pulse length of 16 ns for a π/2 pulse, 32 ns for a π-pulse and 16 ns for the pump pulse. Data points were collected in 8 ns time steps.
2.5 Illumination Protocol
The dark state of ChR2 was prepared by keeping the samples for at least 3 h without any light at 278 K. The light shielded sample was shock frozen in liquid nitrogen, positioned into the precooled resonator (50 K) and DEER traces were recorded. To trap light-activated intermediates of ChR2, the sample was illuminated inside the resonator by a blue LED (470 nm, bandwidth 20 nm (M470F3, Thorlabs)), coupled to a multimode fiber. The light was guided into the resonator using a light fiber sample rod (E5106220, Bruker). The end of the fiber was positioned a few millimeters above the sample volume in the quartz tube. The sample was illuminated for 10 min at 180 K and then allowed to relax at 180 K for 20 min. Then, the resonator was cooled to 50 K and DEER traces were recorded. Similar experiments with the sample illuminated at 180 K and relaxed for 30 min at 205 K, 230 K, or 255 K with subsequent DEER measurements at 50 K were performed. At 278 K a relaxation time of 10 min was applied.
2.6 Analysis of DEER Measurements
The DEER traces were analyzed and validated using DeerAnalysis2018 [35]. The background corrected form factor, F(t), was calculated with a fixed background dimension of 3.0. Application of Tikhonov regularization led to the calculation of the L-curve and the respective distance distributions, P(r). The regularization parameter and the background start value were determined for one DEER trace and fixed within one series of measurements to avoid a bias on the comparison of the distance distributions due to variation of these parameters. In addition, the optimal distance distributions and their errors were calculated using the validation tool of the program. Therefore, the background start (range 232–1232 ns) and density (range 0.3–1.2) were varied and white noise was added to the DEER traces (Lnoise = 1.5). To quantify the observed distance changes, the DEER traces were also fitted with a model-based approach using three Gaussians. The means, the widths, and the relative amplitudes of each Gaussian were allowed to vary during a first round of fittings. For the final fittings of the temperature series, the widths of the two Gaussians with means above 3 nm were held fixed at their average, FWHM = 0.67 nm. The fractional population of each of the three individual Gaussians were calculated by dividing the area for the individual Gaussian by the total area for all Gaussians. The number of interacting spins was calculated from the modulation depths. The value λ = − 0.22 was experimentally determined using a standard for which the number of interacting spins was known to be 2.
2.7 Spin Label Rotamer Analysis and MD Simulations
Interspin distance distributions were simulated applying a rotamer analysis for the spin label side chain and an MD simulation of spin-labeled ChR2 dimer. For the rotamer analysis the program ‘MtsslWizard’ [11], version 2, was used, which calculates 10,000 spin label side chain conformations to find a maximum of 200 rotamers. The clash treatment was set as loose.
MD simulations of spin-labeled ChR2 were carried out in the isobaric-isothermal ensemble (NPT) at 310 K and 1 bar under periodic boundary conditions using the software package ‘YASARA Structure’ [36] with the AMBER14 force field [37]. Particle Mesh Ewald (PME) summation was used for long range electrostatic interactions and a cutoff at 8 Å for van-der-Waals interactions. The time steps for calculation of intra- and intermolecular forces were 1.25 fs and 2.5 fs, respectively. The simulation temperature was controlled by rescaling atom velocities using a Berendsen thermostat [38] based on the time-averaged temperature [39]. Pressure control was achieved by keeping the solvent density at 0.997 g/mL and rescaling the simulation cell isotropic along all three axes. Simulation snapshots were stored each 25 ps. Starting with the ChR2 dimer crystal structure (PDB: 6EID, biological assembly 1), residues C79 and C208 were replaced by the spin label side chain R1 and two independent simulations were performed with this structure. (Only the data for C79 and C79R1 were analyzed in the present work). The spin-labeled ChR2 dimer was inserted into a lipid bilayer composed of POPC/POPG/cholesterol (8:1:1) and placed into a simulation box of 8.7 × 9.4 × 7.0 nm size, filled with TIP3P water and ~ 150 mM sodium and chloride ions, neutralizing the system's net charge. Initial 250 ps equilibration simulations were performed in which the water molecules were restricted from entering the membrane part of the simulation cells to stabilize the bilayer. Afterwards, production runs of 250 ns length were performed, yielding a total simulation time of 0.5 µs.
3 Results
3.1 Interspin Distance Analysis of ChR2–C79R1
For the following experiments, we used a ChR2 mutant that has seven cysteines replaced and only two remaining: C34S/C36S/C87S/C179L/C183L/C208A/C259L (Mut2C) [28]. Spin labeling of Mut2C with native cysteines C79 and C128 using (1-oxyl-2,2,5,5-tetramethyl-pyrroline-3-methyl) methanethiosulfonate spin label (MTSSL) allows the study of interspin distances between helix B sites, C79R1 and C79R1’, in the ChR2 dimer (R1 denotes the spin-labeled side chain). C128 was found to be inaccessible for MTSSL for intact ChR2 [28]. Electrophysiological and spectroscopic experiments supported the conclusion that Mut2C shows wildtype characteristics and the spin-labeled purified proteins did not show any significantly altered kinetics [28]. DEER spectroscopy of spin-labeled Mut2C uncovered light-induced interspin distance changes between positions C79R1 and C79R1’ which were interpreted as movements of helix B upon light activation [28, 29]. A correlation of these movements with the appearance or decay of specific intermediates is still pending.
In the present work, such DEER measurements were extended by an activation and relaxation protocol to investigate freeze-trapped intermediates of Mut2C. Sample illumination was performed with the resonator equipped with a light guide (see Methods) and the sample was illuminated directly inside the resonator. This approach allowed relaxation of the samples at controlled temperatures.
DEER experiments performed with a dark-adapted sample are shown in Fig. 2. Using the model-free Tikhonov regularization to fit the background corrected dipolar evolution function, F(t), the most probable interspin distance between spin labels C79R1 and C79R1’ was found to be 3.7 nm with a shoulder at 4.3 nm. The comparison with distances calculated from the spin label rotamer simulations reveals that the rotamers map a narrower homogeneous distance distribution between these two experimentally found extremes. However, MD simulations (Fig. 3) show that the distances between the spin labels may cluster in three distance regimes centered around 2.3, 3.5 and 4.4 nm (Fig. 2C). Thus, the space sampled by the spin labels in a reconstituted molecular model of ChR2 is larger than that reflected by the rotamer analyses based on a rigid crystal structure. In addition, the occupation of these clusters seems to be correlated with the distances between the corresponding Cα atoms (Fig. 3). Based on this finding and to quantify the distance changes observed upon light activation shown below, we extended our model-free DEER analysis by a model dependent approach using three Gaussians for fitting the DEER traces. The goodness of fit of F(t) to the Gaussian model was similar to that for a model-free Tikhonov analysis (Fig. 2B). Two Gaussians with means at 3.7 and 4.4 nm (FWHM = 0.67 nm) represent the distances found for 83% of the spin pairs and reasonably well reflect the model-free distance distribution between 3 and 5 nm (Fig. 2D). The third Gaussian with a mean at 2.3 nm (FWHM = 1.3 nm) accounts mainly for the probability density below 3 nm. The ratio of the populations with means at 4.4 nm and 3.7 nm amounts to 0.54.
Monitoring the interspin distance between helix B sites, C79R1 and C79R1’, of solubilized ChR2 in the dark state. A Dipolar evolution function, V(t), with background fit. B Form factor, F(t), with fits (smooth lines) applying the model-free Tikhonov regularization or a Gaussian approach. C Distance distribution, P(r), determined from the model-free fits to the data (gray area) with maximum error estimates corresponding to plus/minus two times the standard deviations (dark gray) calculated with the validation tool of DeerAnalysis [35]. The distance distribution calculated from the rotamers (RA, continuous line) and from the two MD simulations shown in Fig. 3 (broken lines) are depicted for comparison. D Distance distribution, P(r), determined from fits of the DEER traces with a sum of three Gaussians (gray area). The populations of the individual Gaussians (lines) are depicted above their maxima
The present result resembles that reported by Sattig et al. [28] and Krause et al. [29]. The distance distribution of Mut2C in the dark-adapted state was reported to show its maximum at 3.7 nm and a shoulder at 4.1 nm [28]. The light-activated state was reported to reveal a distance distribution with the peak and shoulder amplitudes inverted compared to those of the dark state and showed the maximum population at 4.2 nm and the shoulder at 3.8 nm [28]. This light-activated state is evaluated in the next section.
3.2 Trapping and Thermal Relaxation of Light Activated ChR2 States
To correlate the observed movement of helix B with photocycle intermediates, Mut2C was now subjected to a thermal relaxation and trapping protocol (see Sect. 2.5). First, the PK500 state was created by illumination at 180 K followed by thermal relaxation and equilibration at temperatures between 205 and 278 K, after which the sample was cooled again to 50 K for DEER detection. In this way, the thermal energy should allow ChR2 to overcome the energy barrier between its PK500 state and subsequent photocycle intermediates [21]. The corresponding DEER traces and distance distributions are depicted in Fig. 4, validations of the data are shown in the SI, Fig. S1. The distance distributions found for the dark state and the states trapped upon light activation and relaxation at 180 K and 205 K are similar. Thus, the position of helix B is unaltered in ChR2 intermediates trapped up to 205 K in accordance with the TR-SFX experiments on C1C2 [33]. Contrarily, a rise of the probability density for interspin distances at 4.3 nm could be observed upon relaxation of 30 min at 230 K, whose amplitude was further increased upon relaxation at 255 K. Relaxation of the protein for 10 min at 278 K leads back to a distance distribution identical to that found for the dark state.
A The dipolar evolution function, V(t), B the normalized form factor, F(t), with fits, and C the distance distribution, P(r), of freeze-trapped intermediates of solubilized ChR2–C79R1 show temperature dependent changes of the interspin distance distributions. Samples were measured in the dark state, and upon illumination and relaxation at 180 K (20 min), 205 K (30 min), 230 K (30 min), 255 K (30 min), and 278 K (10 min). In (C), for each temperature the distance distribution for the dark state (dashed) is depicted for comparison. Calculations were performed using DeerAnalysis [35] with the background start value and regularization parameter fixed at 232 ns and 100, respectively, to avoid a bias on the comparison due to variation of these parameters. Background fits and validations for the individual DEER measurements with the background start values varied are depicted in Fig. S1 (SI)
The model dependent fittings of the DEER traces using the sum of three Gaussians do not show any significant changes of their mean values (Fig. S2, SI). However, the ratio of the relative amplitudes for the Gaussians centered at 4.4 and 3.6 nm changed from 0.54 for the dark state to 1.38 for the light-activated state relaxed at 255 K. A similar inversion of the height ratios was reported by Sattig et al. [28] for the fully light-activated sample, the shape of the reported distance distribution resembles the present distribution. Thus, the full movement of helix B as observed by DEER occurs in the ChR2 state accumulated at 255 K.
It has been shown that thermal relaxation only allows accumulating the long-lived P480 intermediate [21]. Upon relaxation this state became visible at 217 K with maximum accumulation at 245 K [21]. The increase of the population, P(r), at 4.3–4.4 nm observed here starts to become visible at 230 K with maximum accumulation of this state observed at 255 K. This allows the conclusion that the outward movement of helix B is fully developed in the P480 state. Relaxation at higher temperatures does not show any presence of the P480 state [21], thus, the decrease of the interspin distances observed here is evidence that the relaxation back to the initial state of helix B occurs during the decay of the P480 state.
4 Discussion
The functional importance of the helix B movement is still under discussion. An assignment of this conformational change to specific photocycle intermediates is important to understand its function. The thermal relaxation patterns of the interspin distance changes observed here by DEER are in agreement with the rise and decay of the P480 state as reported in [21]. The photocycle model discussed in [20] includes the P480 state as a possible fourth and last step in the photocycle before the protein relaxes to its initial state. It is populated by up to 25% of the proteins [16, 20]. Identifying the helix B movement to appear during the rise and decay of the P480 state presupposes that structural changes of helix B occur, when the cation channel is already closed and are only performed by a small fraction of the protein when the illumination time is short. A functional model with two closed and two open states was introduced in 2009 [40]. A recently published unifying photocycle model [17] discriminates between two photocycles, the anti-cycle, which is responsible for the channel conductivity for cations and protons, and the syn-cycle, which characterizes the opening and closing process for only protons. In this model, the P480 state as part of the syn-cycle is accumulated during continuous illumination. The authors identified the deprotonation of E90 to occur only in the syn-cycle, which was confirmed to occur in the submicrosecond time range [27] and which has to be related to the rise of the P480 state. In the present work, this rise is suggested to be accompanied by the outward movement of helix B. It was further shown that the relaxation of the P480 state to the ground state is correlated with the rearrangement of the helical backbone of ChR2 [15]. Our data show that this rearrangement includes the movement of helix B back to the initial state.
The present results provide evidence that the movement of helix B might work as or is indication of a switch between the anti- and the syn-cycle as suggested in [8]. In this case, it would be responsible for desensitizing the channel for cation conduction. As the accumulation of the P480 state is achieved by continuous illumination, the conductivity of the channel is adapted depending on illumination time. Thereby, the flux of cations is controlled directly by the protein, when the illumination time becomes longer. However, our experiments cannot exclude that the movement of helix B already occurs in the PN520 state in accordance with results on the ChR2–C128T mutant [29] and is thus also important for ion permeation and selectivity.
In summary, we demonstrated that a light-induced movement of helix B is correlated with the presence of the P480 state of ChR2. We hypothesize that conformational changes occurring in this area are key elements responsible for desensitizing the channel for cation conduction.
Availability of Data and Material
All data generated or analyzed during this study are included in this published article.
Code Availability
Not applicable.
References
O. Yizhar, L.E. Fenno, T.J. Davidson, M. Mogri, K. Deisseroth, Neuron 71, 9 (2011)
G. Nagel, T. Szellas, W. Huhn, S. Kateriya, N. Adeishvili, P. Berthold, D. Ollig, P. Hegemann, E. Bamberg, Proc Natl Acad Sci USA 100, 13940 (2003)
J.P. Klare, I. Chizhov, M. Engelhard, Bioenergetics (2007). https://doi.org/10.1007/400_2007_041
J. Pinhassi, E.F. DeLong, O. Béjà, J.M. González, C. Pedrós-Alió, Microbiol Mol Biol Rev 80, 929 (2016)
M. Muller, C. Bamann, E. Bamberg, W. Kuhlbrandt, J Mol Biol 414, 86 (2011)
H.E. Kato, F. Zhang, O. Yizhar, C. Ramakrishnan, T. Nishizawa, K. Hirata, J. Ito, Y. Aita, T. Tsukazaki, S. Hayashi, P. Hegemann, A.D. Maturana, R. Ishitani, K. Deisseroth, O. Nureki, Nature 482, 369 (2012)
O. Volkov, K. Kovalev, V. Polovinkin, V. Borshchevskiy, C. Bamann, R. Astashkin, E. Marin, A. Popov, T. Balandin, D. Willbold, G. Buldt, E. Bamberg, V. Gordeliy, Science 358, eaan8862 (2017)
R. Richards, R.E. Dempski, J Biol Chem 292, 7314 (2017)
C. Bamann, R. Gueta, S. Kleinlogel, G. Nagel, E. Bamberg, Biochemistry 49, 267 (2010)
M. Nack, I. Radu, M. Gossing, C. Bamann, E. Bamberg, G.F. von Mollard, J. Heberle, Photochem Photobiol Sci 9, 194 (2010)
G. Hagelueken, R. Ward, J.H. Naismith, O. Schiemann, Appl Magn Reson 42, 377 (2012)
J.L. Spudich, C.-S. Yang, K.-H. Jung, E.N. Spudich, Annu Rev Cell Dev Biol 16, 365 (2000)
C. Bamann, T. Kirsch, G. Nagel, E. Bamberg, J Mol Biol 375, 686 (2008)
E. Ritter, K. Stehfest, A. Berndt, P. Hegemann, F.J. Bartl, J Biol Chem 283, 35033 (2008)
I. Radu, C. Bamann, M. Nack, G. Nagel, E. Bamberg, J. Heberle, J Am Chem Soc 131, 7313 (2009)
V.A. Lorenz-Fonfria, T. Resler, N. Krause, M. Nack, M. Gossing, G. Fischer von Mollard, C. Bamann, E. Bamberg, R. Schlesinger, J. Heberle, Proc Natl Acad Sci USA 110, E1273 (2013)
J. Kuhne, J. Vierock, S.A. Tennigkeit, M.A. Dreier, J. Wietek, D. Petersen, K. Gavriljuk, S.F. El-Mashtoly, P. Hegemann, K. Gerwert, Proc Natl Acad Sci U S A 116, 9380 (2019)
V.A. Lorenz-Fonfria, J. Heberle, Biochim Biophys Acta 1837, 626 (2014)
S. Bruun, D. Stoeppler, A. Keidel, U. Kuhlmann, M. Luck, A. Diehl, M.A. Geiger, D. Woodmansee, D. Trauner, P. Hegemann, H. Oschkinat, P. Hildebrandt, K. Stehfest, Biochemistry 54, 5389 (2015)
M. Saita, F. Pranga-Sellnau, T. Resler, R. Schlesinger, J. Heberle, V.A. Lorenz-Fonfria, J Am Chem Soc 140, 9899 (2018)
J. Becker-Baldus, C. Bamann, K. Saxena, H. Gustmann, L.J. Brown, R.C. Brown, C. Reiter, E. Bamberg, J. Wachtveitl, H. Schwalbe, C. Glaubitz, Proc Natl Acad Sci USA 112, 9896 (2015)
H.-J. Steinhoff, R. Mollaaghababa, C. Altenbach, K. Hideg, M. Krebs, H.G. Khorana, W.L. Hubbell, Science 266, 105 (1994)
H.J. Sass, G. Büldt, R. Gessenich, D. Hehn, D. Neff, R. Schlesinger, J. Berendzen, P. Ormos, Nature 406, 649 (2000)
J.P. Klare, E. Bordignon, M. Engelhard, H.-J. Steinhoff, Photochem Photobiol Sci 3, 543 (2004)
J.L. Spudich, Mol Microbiol 28, 1051 (1998)
T. Nakanishi, S. Kanada, M. Murakami, K. Ihara, T. Kouyama, Biophys J 104, 377 (2013)
J. Kuhne, K. Eisenhauer, E. Ritter, P. Hegemann, K. Gerwert, F. Bartl, Angew Chem Int Ed Engl 54, 4953 (2015)
T. Sattig, C. Rickert, E. Bamberg, H.J. Steinhoff, C. Bamann, Angew Chem Int Ed Engl 52, 9705 (2013)
N. Krause, C. Engelhard, J. Heberle, R. Schlesinger, R. Bittl, FEBS Lett 587, 3309 (2013)
H. Li, O.A. Sineshchekov, G. Wu, J.L. Spudich, J Biol Chem 291, 25319 (2016)
M. Muller, C. Bamann, E. Bamberg, W. Kuhlbrandt, J Mol Biol 427, 341 (2015)
M. Takemoto, H.E. Kato, M. Koyama, J. Ito, M. Kamiya, S. Hayashi, A.D. Maturana, K. Deisseroth, R. Ishitani, O. Nureki, PLoS ONE 10, e0131094 (2015)
K. Oda, T. Nomura, T. Nakane, K. Yamashita, K. Inoue, S. Ito, J. Vierock, K. Hirata, A.D. Maturana, K. Katayama, Elife 10, e62389 (2021)
N. Krause, Structural rearrangements upon opening of Channelrhodopsin-2. Refubium - Repositorium der Freien Universität Berlin (2016). https://doi.org/10.17169/refubium-13743
G. Jeschke, V. Chechik, P. Ionita, A. Godt, H. Zimmermann, J. Banham, C. Timmel, D. Hilger, H. Jung, Appl Magn Reson 30, 473 (2006)
E. Krieger, G. Koraimann, G. Vriend, Proteins: Struct Funct Bioinform 47, 393 (2002)
J.W. Ponder, D.A. Case, Adv Protein Chem 66, 27 (2003)
H.J. Berendsen, Jv. Postma, W.F. van Gunsteren, A. DiNola, J.R. Haak, J Chem Phys 81, 3684 (1984)
E. Krieger, T. Darden, S.B. Nabuurs, A. Finkelstein, G. Vriend, Proteins: Struct Funct Bioinform 57, 678 (2004)
K. Nikolic, N. Grossman, M.S. Grubb, J. Burrone, C. Toumazou, P. Degenaar, Photochem Photobiol 85, 400 (2009)
Acknowledgements
We thank Ernst Bamberg (MPI of Biophysics) for continuous support and Heike Biehl for help with the ChR2 sample preparation.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was financially supported by the Max Planck Society to CB and the Deutsche Forschungsgemeinschaft to HJS (DFG, SFB 944) and to CB (DFG, SFB 807-P12).
Author information
Authors and Affiliations
Contributions
Conceptualization: CB, HJS; methodology: MS, JPK, CB, HJS; formal analysis and investigation: MS, HJS; writing—original draft preparation: MS; writing—review and editing: MS, CB, JPK, HJS; funding acquisition: CB, HJS; resources: CB, HJS.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schumacher, M., Klare, J.P., Bamann, C. et al. DEER Spectroscopy of Channelrhodopsin-2 Helix B Movements in Trapped Photocycle Intermediates. Appl Magn Reson 53, 731–743 (2022). https://doi.org/10.1007/s00723-021-01380-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00723-021-01380-9