Abstract
Porcine circovirus 2 (PCV2) is associated with a series of swine diseases. There is a great interest in improving our understanding of the immunology of PCV2, especially the properties of the viral capsid protein Cap-PCV2 and how they relate to the immunogenicity of the virus and the subsequent development of vaccines. Phage display screening has been widely used to study binding affinities for target proteins. The aim of this study was to use phage display screening to identify antigenic peptides in the PCV2 capsid protein. After the selection of peptides, five of them presented similarity to sequences found in cap-PCV2, and four peptides were synthesized and used for immunization in mice: 51–CTFGYTIKRTVT-62 (PS14), 127-CDNFVTKATALTY-138 (PS34), 164-CKPVLDSTIDY-173 (PC12), and 79-CFLPPGGGSNT-88 (PF1). Inoculation with the PC12 peptide led to the highest production of antibodies. Furthermore, we used the PC12 peptide as an antigen to examine the humoral response of swine serum by ELISA. The sensitivity and specificity of this assay was 88.9% and 92.85%, respectively. Altogether, characterization of immunogenic epitopes in the capsid protein of PCV2 may contribute to the improvement of vaccines and diagnostics.
Similar content being viewed by others
Introduction
Porcine circovirus 2 (PCV2) is one of the smallest viruses that infects animal cells, and is comprised of a non-enveloped capsid about 23 nm in diameter [1]. Taxonomically, PCV2 is classified within the Circoviridae family, genus Circovirus, and is associated with a number of diseases in pigs, called porcine circovirus associated diseases (PCVAD) [2]. Postweaning multisystemic wasting syndrome (PMWS) has become the main syndrome associated with PCVAD, and has been diagnosed in pigs in Asia, South and North America, and Europe. Due to the large worldwide spread and economic losses generated by PCVAD, PCV2 is one of the most important pathogens to the global swine industry [3].
Understanding the immune response and antigenicity of the PCV2 viral particle during infection contributes to the development of more efficient strategies to control this pathogen [4]. Currently, the main approach to control PCVD is through vaccination of animals [5].
Vaccines against PCV2 generate an immune response against the viral capsid, a supramolecular structure formed by twelve pentamers of a single protein called Cap [6]. This protein has a primary structure consisting of 233 (or 234) amino acid residues with a molecular mass of approximately 28 kDa [7]. Structurally, it has two β-sheets in which one is composed of four antiparallel β-strands. These strands are connected by four short (4 to 9 amino acid residues) and three long loops (21 to 36 amino acid residues) [8].
Phage display (Ph.D.) screening has been used to study binding affinities for protein targets [9,10,11,12,13,14]. Different techniques have been used to identify epitopes in Cap-PCV2 [15,16,17]. Ge et al. [18], identified three new antigenic regions in Cap-PCV2 using Ph.D. screening. Because this technique can employ different libraries and strategies of selection, it makes the identification of new epitopes in the Cap protein feasible. To this end, we first utilized Ph.D. screening to identify immunogenic sequences in Cap-PCV2. Second, the IgG1 and IgG2a antibody responses of mice inoculated with the immunogenic peptides were evaluated. Finally, we used the PC12 peptide, identified by Ph.D. screening, as a tool for the serological diagnosis of PCV2-infected pigs.
Materials and methods
Purification of Cap-PCV2
The DNA coding sequence of Cap-PCV2 (GenBank: ABD42929) was cloned into the pET-16b vector (Novagen), according to Salgado et al. [19]. The N-terminus 6x Histidine tagged-viral protein was expressed in Escherichia coli strain BL21-CodonPlus (DE3) - RIL (Agilent Technologies). Protein purification was performed by affinity chromatography using a HisTrap crude FF (GE), according to the manufacturer’s recommendations in a FPLC system (AKTA TM purifier). The purity of the recombinant protein was checked in a 12% SDS-PAGE gel.
Selection of scFv clones
A human scFv phage library containing 2x106 antibody fragment sequences [20] was used to select antibodies against the recombinant and purified Cap-PCV2. Two selection rounds were carried out to amplify the scFv library in E. coli strain XL1 Blue (Stratagene) with the concomitant infection of the helper VCSM13 phage [21]. In total, each well in a 96-well microplate (Nunc) was sensitized with 4 μg of Cap-PCV2 in 0.1 M bicarbonate buffer (pH 9.6), and incubated for 14 hours at 4°C. The blocking step was performed in phosphate buffer saline (PBS) containing 3% bovine serum albumin (BSA) for 1 hour at 37°C. Subsequently, 50 μL of each phage solution was added to an individual well and the plate was incubated for 2 hours at 37°C. The plate was then washed 10 times with PBS containing 0.005% (v/v) Tween 20 (PBS-T). Finally, phages were eluted in 50 μL of 0.1 M HCl solution (pH 2.2), and subsequently neutralized with 2 M of Tris-HCl buffer (pH 9.0).
Induction of scFv peptides expression
Plasmids were isolated from previously selected E. coli strain XL1 Blue and used to transform E. coli strain Top 10 (Invitrogen) competent cells [21]. Bacterial cells were then plated in a solid Luria-Bertani (LB) medium containing 100 μg.ml−1 carbenicillin. After 12 hours of incubation at 37°C, transformed colonies were transferred to a deep 96-well plate containing Super Broth (SB) medium supplemented with 2M glucose (v/v) and 100 ug.ml−1 carbenicillin. The plate was then incubated for 12 hours at 37°C and 250 rpm. After centrifugation at 2250 g for 10 minutes, supernatants were discarded and pellets were resuspended in SB medium containing 2.5 mM isopropyl β-D-thiogalactopyranoside inducer (IPTG) (Sigma), 100 μg.ml−1 carbenicillin and 2 M glucose. The plate was later incubated for another 12 hours at 30°C and 250 rpm. Finally, the plate was centrifuged at 4000 g for 10 minutes at 4°C and supernatants were collected for analysis of expression of scFv clones.
Analyses of protein expression and specificity of scFv clones
To analyze protein expression in the scFv clones, 10 μL of the supernatants derived from scFv clones diluted in 40 μL bicarbonate buffer (pH 9.6) were individually immobilized on microtiter wells of a MaxiSorp™ plate (Nunc). To determine the specificity of clones, each well of a MaxiSorp™ plate (Nunc, Denmark) was sensitized with 0.8 μg of Cap-PCV2 or BSA (negative control), both diluted in bicarbonate buffer, followed by addition of scFv clones induced supernatants into each well. In both analyses, we used a peroxidase conjugated anti-HA antibody (1:1000) (Roche, Switzerland). Color development was achieved by adding a solution containing H2O2, orthophenylenodiamine (OPD), and 0.1 M citrate buffer (pH 5.0). Readout was performed at a wavelength of 492 nm by an Enzyme-Linked Immunosorbent Assay (ELISA) plate reader.
Western Blotting
Purified 6x Histidine tagged-Cap-PCV2 was subjected to a 12% SDS-PAGE gel, and after electrophoresis proteins were transferred to a nitrocellulose membrane (GE) using a Mini Trans-Blot® electrophoretic transfer (Bio-Rad) according to the manufacturer’s instructions. The membrane was first incubated with blocking buffer for 1 hour at room temperature. Afterwards, the membrane was washed with PBS-T and then cut in strips which were incubated for 1 hour in 3 mL of F1 and F5 scFv clones supernatants, commercial anti-PCV2 polyclonal antibody (VMRD) (positive control) or PBS (negative control). After washing three times with PBS-T, the respective antibodies were added: anti-HA peroxidase conjugated antibody (Roche, Switzerland) (1:2000) to the negative control, F1 and F5 scFv clones; and anti-swine IgG peroxidase conjugated secondary antibody (Sigma) (1:10,000) to the positive control. Immunoreactive protein bands were revealed with 3.3 tetrahydrochloride (DAB) and 0.015% (v/v) hydrogen peroxide in PBS.
Selection of Cap-PCV2 mimetic peptides
We performed the phage display screening using anti-PCV2 polyclonal antibodies (VMRD), an in-house rabbit anti-PCV2 polyclonal antibody purified by the caprylic acid precipitation method [22], and supernatants derived from F1 and F5 scFv clones.
Phage display libraries, obtained from New England Biolabs, Inc., Beverly, MA,
contained peptide sequences (7-mer or 12-mer) inserted in the pIII minor coat protein N-terminus of the M13 bacteriophage. The Ph.D.-C7C and the Ph.D.-12 libraries consisted of approximate 1.2x109 and 1.9x109 independent clones, respectively. Ph.D.-C7C and Ph.D.-12 libraries were screened through biopanning in a solution containing either protein G magnetic beads (Invitrogem) to retrieve the antibody-phage complex or nickel charged magnetic agarose beads (Qiagen) to retrieve 6x Histidine tagged-scFv fragments.
Biopannings were performed with three rounds of selection when phages were screened using swine serum, and two rounds of selection when phages were screened with scFv fragments. A combination of 1x1011 viral particles of Ph.D.-12 with either 4x108 beads/mL-antibody (swine serum) or beads-scFv were incubated for 1 hour at 37°C. After incubation, beads were washed 10 times with PBS-T and the unbound phage particles were discarded, followed by competitive elution of bound phages with 50 µg of purified Cap-PCV2 for 10 minutes at room temperature. Eluted phages were amplified in E. coli strain ER2738 (New England Biolabs) and purified using PEG-NaCl precipitation. After each round of biopanning, isolated bacterial colonies containing amplified phage clones were grown in a microtiter plate and titrated essentially as described by Barbas III et al. [21].
The biopanning using rabbit polyclonal antibodies was performed in two steps. The first step consisted of a subtractive step to remove phages that bound non-specifically on the magnetic beads. The second step consisted of retrieving those phages that expressed peptides bound to antibodies against Cap-PCV2. The removal of non-specific phages, a combination of 1x1011 viral particles of Ph.D.-C7C with 4x108 beads/mL-antibody derived from the negative serum for PCV2 was performed. After 1 hour of incubation at 37°C, the supernatant was used for positive selection, which was performed with microspheres bound to positive rabbit serum IgG for PCV2 antigen, as previously done for Ph.D.-12.
Sequencing and in silico analysis
The DyEnamic ET Dye Terminator Cycle Sequencing Kit (GE Healthcare) and a MegaBaceTM sequencer 1000 (GE Healthcare) were used to sequence the heavy and light chains of the scFv clones previously selected by ELISA using Cap-PCV2 as the coating protein. Phage DNA sequences encoding the recombinant peptides selected from the Ph.D. were sequenced. The primers used were the following: MMB4 (5’-TCC TCC GCT GGC TAT GTG GTT T-3’) for the light chain, MMB5 (5’-CGT TTG CCA TTT CAT AAT TCT C-3’) for the heavy chain of scFv clones, and -96 M13 (5’-CCCTCATTAGTTAGCGCGTAACG-3’) for the phage display peptides. The scFv nucleotide sequences were analyzed using IgBlast (http://www.ncbi.nlm.nih.gov/igblast). The peptides sequences were aligned with Cap-PCV2 amino acid sequence using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins). The three dimensional structures of the Cap-PCV2 found in the Protein Data Bank (PDB), e.g. 3R0R, and the PCV2 capsid sequence were used for structural inferences using the program PyMOL (DeLano Scientific).
Phage-ELISA
PoliSorp microplates (Nunc) were coated with each clone at a concentration of 5.0x1010 phages/mL, previously diluted in bicarbonate buffer, pH 9.6, and incubated at 4°C for 14 hours. Plates were blocked for 1 hour at 37°C with 10% (w/v) non-fat dry milk in PBS, followed by three washes with PBS. Positive or negative rabbit antibodies for PCV2 were previously incubated with M13 phage (wild-type) (1011 phages/mL) and then added to microplates at 1:400 dilution in blocking solution and incubated for 1 hour at 37°C. After washing three times with PBS, a commercial anti-rabbit IgG conjugated to peroxidase (Sigma) at 1:2000 dilution in blocking solution was added to each well. Color development was achieved by adding a solution containing H2O2, OPD, and 0.1 M citrate buffer (pH 5.0). Readout was performed at a wavelength of 492 nm. Wild-type M13 phage was used as a negative control. Each clone was tested in duplicate.
Synthetic Peptides
Four synthetic peptides (PS14, PS34, PF1, and PC12) (Table. 1) were designed from the selected phages and they were chemically synthesized and conjugated to Keyhole Limpet Hemocyanin (KLH) using PEPTIDE 2.0 (Peptide 2.0 Inc., USA).
Immunizations
All experimental procedures were conducted in compliance with the ethical principles of the Brazilian Academy of Animal Experimentation and approved by the Animal Research Ethics Committee of the Universidade Federal de Viçosa under the protocol number 39/2012.
The two following immunization experiments were conducted in mice: (1) using selected phages as antigens, and (2) using the conjugated synthetic peptides as antigens.
Experiments were carried out in 4–6 weeks old male BALB/c mice. In the first trial, mice were divided into 8 groups of 5 mice each. Immunizations were performed with three subcutaneous injections with intervals of 15 days for each and using the following phages: S14, S34, C12, FV7, and F1. Each inoculum contained 1010 phages and 25% aluminum hydroxide as an adjuvant. The negative control inoculums contained PBS and wild-type M13 phage. The positive control contained Cap-PCV2 recombinant protein. In the second trial, mice were divided into 6 groups of 6 mice each. Immunizations were performed with three subcutaneous injections with intervals of 15 days for each and using the following peptides, 100 µg per dose: PS14, PS34, PC12, and PF1 conjugated to KLH. The negative and positive controls contained PBS and purified cap-PCV2 protein, respectively. Saponin (Sigma) at 100 μg per dose was used as an adjuvant in combination with the peptides and the controls. Blood samples were collected 45 days after immunization with phages, and at 0, 15, 30, and 45 days after immunization with peptides. Sera were later examined for the presence of specific antibodies.
Detection of IgG, IgG1, and IgG2a antibodies for Cap-PCV2
A 96-well Maxisorp microtiter plate (Nunc) was coated with 0.36 ng/well of Cap-PCV2 diluted in bicarbonate buffer. After incubation for 14 hours at 4°C, wells were blocked with 5% BSA in PBS. Sera from animals inoculated with either phage or peptides were diluted in blocking buffer at 1:20 and 1:50, respectively, before being added to the plate, followed by incubation at 37°C for 1 h. After washes, peroxidase-conjugated anti-mouse IgG, IgG1, or IgG2a antibodies (Sigma), diluted at 1:5000, were added to the plate and incubated for 1 hour at 37°C. Color development was achieved by adding a solution containing H2O2, OPD, and 0.1M citrate buffer (pH 5.0). Readout was performed at a wavelength of 492 nm. Each sample was tested in triplicate.
Evaluation of swine serum response by PC12 peptide based ELISA
To investigate whether the PC12 peptide can be used as a serological tool for detecting PCV2 infection, twenty-seven positive swine serum and 14 negative swine serum samples which had been validated by the peroxidase monolayer assay (IPMA) were used for the evaluation [23]. In addition, 20 swine which had not been vaccinated in the field were naturally infected with PCV2, and swine sera were collected for evaluation at 52, 102, and 166 days old. To conduct in house ELISA, 5 ug/well of PC12 or PCV2-Cap in bicarbonate buffer was immobilized on a 96-well Maxisorp microtiter plate (Nunc). After incubation for 14 hours at 4°C, wells were blocked with 5% BSA in PBS. Sera from swine were diluted in blocking buffer at 1:50 and then added to the plate for 1 h at 37°C. After washes, peroxidase-conjugated anti-pig IgG antibodies (Sigma), diluted at 1:5000, were added to the plate and incubated for 1 hour at 37°C. Color development was achieved by adding a substrate solution containing H2O2, OPD, and 0.1 M citrate buffer (pH 5.0). Readout was performed at a wavelength of 492 nm by an ELISA plate reader (company name). Each sample was tested in triplicate.
Statistical analyses
Differences between groups were analyzed using ANOVA test, followed by the Dunnett’s test. Statistical analyses were done using GraphPad Prism 5.0 (GraphPad Software Inc., San Diego, CA). P values that were less than 0.05 were considered statistically significant.
Results
Peptide selection using scFvs clones
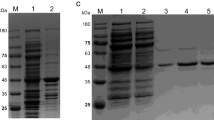
After selection of scFv clones, the expression and affinity of fragments against Cap-PCV2 were assessed. All scFv fragments that were expressed and exhibited absorbance above 0.2, as detected by ELISA (Fig. S1), were tested for their capacity to bind to cap-PCV2. Most scFv fragments presented a high affinity for Cap-PCV2 (Fig. S2A). F1 and F5 fragments showed higher levels of expression and binding affinity to Cap-PCV2 when compared to the others. Therefore, the F1 and F5 clones were selected to perform a Ph.D. screen using the phage libraries for the peptides. The binding capacity of F1 and F5 scFv fragments towards Cap-PCV2 was confirmed by Western blotting (Fig. S2B). Cloned nucleotide sequences were analyzed by IgBlast to determine the complementarity determining regions (CDRs) (Fig. S2C).
We used F1 and F5 scFv clones for phage selection of the target peptide and found 8 and 6 clones using F1 and F5 scFv antibodies, respectively (Table S1). The FV7 and F1 peptides showed some sequence similarity to the cap-PCV2 protein (Table 1). The peptide FV7 is located towards the inside of the predicted capsid structure (Fig. 1).
Peptide selection using PCV2 capsid-specific polyclonal antibodies
A subtractive step was performed in the biopanning using in house negative serum for Cap-PCV2 to improve the affinity and specificity of the in house polyclonal antibody for Cap-PCV2 (positive serum). Another selection strategy included a binding step with anti-PCV2 pig commercial serum to the target peptides. Titration results suggested that our selection strategies were efficient since there was an enrichment of specific phages over the selection cycles (data not shown).
After sequencing of clones, the selected peptides were aligned with the Cap-PCV2 amino acid sequence (Table. 1 and Table. S1). There were no cleavage sites on the Cap-PCV2 sequence used for the alignment (aiming to cover both PCV2a and PCV2b sequences).
The selected peptides located on the capsid surface, including the peptides S14 and S34. One peptide (C12) was located at a junction region of Cap-PCV2 monomers but is still on the outside portion of the capsid structure (Fig. 1).
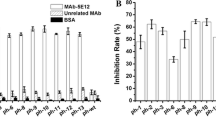
Evaluation of selected phages with anti-PCV2 polyclonal antibody
The reactivity of phages which showed the best peptide alignments with Cap-PCV2 sequences was evaluated by ELISA using in house negative and positive sera for PCV2. All the clones tested showed a higher reactivity with the positive serum when compared to the negative serum (Fig. 2), confirming the affinity of these phages towards anti-Cap-PCV2 antibodies.
Differential IgG production in mice immunized with selected phages and peptides
Our results showed that mice inoculated with F1, FV7, S14, or S34 phages produced antibodies against Cap-PCV2 15 days after the third immunization (day 45). On the other hand, the group inoculated with the C12 phage showed no statistical difference when compared to the group immunized with the wild-type phage (Fig. 3).
Anti Cap-PCV2 IgG mouse production using selected phages as vaccine candidates. Mice were immunized three times with 1010 phages/mL. Fifteen days after the last inoculation each serum was tested by ELISA. WT: wild-type M13 phage. S/P ratio = (OD sample - OD negative control)/(OD positive control - OD negative control). Asterisks indicate differences statistically significant (P < 0.05)
Mice inoculated with synthetic peptides showed differing IgG antibody production patterns. For instance, mice immunized with the PC12 peptide showed a higher production of total IgG (Fig. 4A) and IgG1 (Fig. 4B) antibodies than the control group 15 days after the third inoculation while mice immunized with the other peptides showed no statistical difference in the IgG antibody production when compared to the control group. In addition, the IgG2a antibody production was higher in mice inoculated with the PF1 peptide when compared to the control while the other peptides tested did not induce a significant IgG2a antibody production (Fig. 4C). Mice inoculated with the purified Cap-PCV2 showed the highest antibody production for both IgG isotypes when compared to the control group.
Production of anti-Cap-PCV2 IgG isotypes before (day 0) and after immunization (days 15, 30, and 45) with synthetic peptides. A) total IgG; B) IgG1; C) IgG2a. S/P ratio; (OD sample - OD negative control)/(OD positive control - OD negative control). Asterisks indicate differences statically significant (P < 0.05)
Reactivity of pig serum against the PC12 peptide
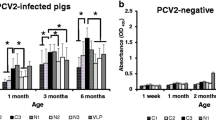
The PC12 peptide was able to discriminate sera from animals infected with PCV2 from negative animal sera showing a sensitivity of 88.9% and specificity of 92.85% (Fig 5A).
Detection of PCV2 positive sera from the field by PC-12 peptide based ELISA. A) Positive sera against PCV2 (n = 29) and negative control sera (n = 14) were evaluated for PC12 reactivity by ELISA (cut-off 0.1). B) Investigation of a humoral response in PCV2-infected pigs at different time points, using PC-12 peptide and Cap-PCV2 based ELISA. S/P ratio; (OD sample - OD negative control)/(OD positive control - OD negative control)
We examined animal serum from different production periods on the same farm. The animals were not vaccinated against PCV2 and the animals were naturally challenged (against PCV2). In addition all animals were positive for PCV2-DNA (data not shown). The animals from these different periods presented a serological profile against PC12 similar to their serological profile against the complete capsid protein antigen (Fig 5B).
Discussion
The selection of antigenic mimetic peptides by Ph.D. screening has been successfully used with high accuracy [24]. This technique allows monoclonal or polyclonal antibodies to bind to selected peptides from a phage display library. This research used phage display screening to identify antigenic regions in the Cap protein from PCV2 and found four peptides in the capsid protein, as summarized in Fig 6. The antigenic regions were assessed by ELISA as well as in immunization trials with either selected phages or synthetic peptides in mice. In addition, we investigated serology response to the PC12 peptide in pigs naturally infected with PCV2.
Vaccination against PCV2 is an important measure to prevent PCVD [25, 26]. All vaccines currently on the market are based on Cap-PCV2 immunogenicity, and are either recombinant or contain inactivated virus. The immune response against PCV2 is generated by a balance between humoral and cell-mediated immune responses. The latter is not completely understood, although it has been demonstrated that Cap-PCV2 and the protein encoded by ORF 1 (Rep) are involved in this response [27]. Furthermore, CD4+ and CD8+ cells and IFN-γ secreting cells seem to play a role during the cell-mediated immune response [28, 29]. The humoral response is responsible for inducing the production of neutralizing antibodies [30]. Meerts et al. [31] demonstrated that pigs affected by PMWS have lower titers of neutralizing antibodies when compared to healthy animals. Additionally, Fort et al. [32] postulated that levels of neutralizing antibodies are related to the clinical presentations seen in the animal. These findings highlight the importance of understanding the antigenic and immunogenic regions of the Cap-PCV2 protein.
Mice inoculated with selected phages and synthetic peptides showed differing production of antibody against Cap-PCV2. Animals inoculated with the recombinant phages S14, S34, F1, and FV7 showed a higher production of total IgG antibody against Cap-PCV2 than those inoculated with the wild-type phage. Alternatively, the production of total IgG against Cap-PCV2 in mice immunized with the synthetic peptides PS14, PS34, and PF1 showed no statistical difference when compared to the control group. PC12 was the only synthetic peptide that stimulated the production of total IgG antibody against Cap-PCV2 when compared to the control group. The differences observed in antibody production when inoculation was performed with phages, versus synthetic peptides, might be explained by the fact that phages can act as an adjuvant, enhancing the humoral response [33].
Our study also evaluated the production of IgG1 and IgG2a isotypes in mice immunized with the selected phages and synthetic peptides. It is generally recognized that, in mice, the cell-mediated immune response induced by Th1 cells is associated with the production of IgG2a, whereas the humoral response induced by Th2 cells is associated with the production of IgG1 [34]. Our results showed that mice immunized with purified Cap-PCV2 presented a higher concentration of both IgG1 and IgG2a isotypes than any other phage or synthetic peptide tested. Among the four synthetic peptides tested, PC12 induced the highest production of IgG1 isotype. In addition, PC12 showed reactivity with serum from pigs naturally infected with PCV2.
The antigenic region (PC12) found in this experiment is located at a junction region between Cap-PCV2 monomers. However, PC12 is present on the outside face of the virion. Mahe et al. [15] and Lekcharoensuk et al. [16] demonstrated that the regions 169–183 and 165–200 are antigenic regions, respectively. Since PC12’s residues (168–173) are within these two previously described antigenic regions, it is feasible that PC12 actually contains the antigenic epitope. Of note, PC12 represents a smaller antigenic site than the results from previous studies [15, 16].
The production of IgG2a isotype antibody was statistically different in the group of mice immunized with PF1. This synthetic peptide was derived from a region covering amino acid residues 79–88 present in the Cap-PCV2 sequence. A previous study showed that the amino acid residues 65–87 are potentially immunogenic [15], which is consistent with our findings. Additionally, because this region is associated with the production of IgG2a isotype, it might also be involved in the Th1 response during PCV2 infection. The PF1 antigenic site could be used for further investigation of the Th1 immune response in pigs in the future.
To summarize previous studies that have identified immunogenic regions in the Cap-PCV2 protein, (1) Mahe et al. [15] used synthetic peptides and pig sera obtained from animals inoculated with isolated PCV2 to detect the presence of antibody binding regions among the amino acid residues 25–47, 65–87, 113–147, 169–183, and 193–207. (2) Lou et al. [35] reported the existence of an immunodominant region among the amino acid residues 122–136. (3) A study using a set of monoclonal antibodies and chimeric PCV1–PCV2 constructs identify three different immunogenic regions among the amino acid residues 47–63, 165–200, and 200–233 [16]. (4) Shang et al. [17] reported the existence of four epitope regions recognized by monoclonal antibodies, and those regions were found among the amino acid residues 156–162, 175–192, 195–202, and 231–233. (5) Ge et al. [18] reported the following antigenic regions: 50–56, 62–67, 68–73, 68–73, 79–84, 86–93, 102–107, 119–128, and 229–233.
Phage display screening has previously been used for epitope mapping in Cap-PCV2 by Ge et al. [18], but here we used different phage display libraries. Our analysis showed four different epitope regions on Cap-PCV2, which included amino acids 52–56, 83–88, 130–142, and 168–173. As shown in Fig. 6, our results corroborate previous epitope mapping studies.
Here, we identified smaller antigenic regions than those identified in previous studies. Additionally, the PF1 and PC12 antigenic sites showed serological responses in a mouse model. Further, PC12 was used as an antigen in an ELISA assay with pig serum, with these results validating that PC12 displayed an important antigenic site relevant during PCV2 infection. The PC12 region might be of use as a target for the future design of new recombinant antigens.
In conclusion, we discovered Cap-PCV2 mimetic peptides capable of inducing antibody production against Cap-PCV2 when inoculated in mice. We also found that PC12 (CKPVLDSTIDY) could serve as an antigenic site in pigs infected with PCV2. Therefore, our data further elucidates the immunogenic regions of Cap-PCV2, which may ultimately improve vaccine development.
References
Crowther RA, Berriman JA, Curran WL et al (2003) Comparison of the structures of three circoviruses: chicken anemia virus, porcinecircovirus type 2, and beakand feather diseasevirus. J Virol 77:13036–13041. https://doi.org/10.1128/JVI.77.24.13036
Segalés J, Allan GM, Domingo M (2005) Porcine circovirus diseases. Anim Heal Res Rev 6:119–142. https://doi.org/10.1079/AHR2005106
Segalés J, Kekarainen T, Cortey M (2013) The natural history of porcine circovirus type 2: from an inoffensive virus to a devastating swine disease? Vet Microbiol 165:13–20. https://doi.org/10.1016/j.vetmic.2012.12.033
Darwich L, Mateu E (2012) Immunology of porcine circovirus type 2 (PCV2). Virus Res 164:61–67. https://doi.org/10.1016/j.virusres.2011.12.003
Segalés J (2015) Expert REVIEW OF VACCINES BEST practice and future challenges for vaccination against porcine circovirus type 2. Expert Rev Vacc 14:473–487. https://doi.org/10.1586/14760584.2015.983084
Blanchard P, Mahé D, Cariolet R et al (2003) Protection of swine against post-weaning multisystemic wasting syndrome (PMWS) by porcine circovirus type 2 (PCV2) proteins. Vaccine 21:4565–4575. https://doi.org/10.1016/S0264-410X(03)00503-6
Hamel AL, Lin LL, Nayar GP (1998) Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J Virol 72:5262–5267
Khayat R, Brunn N, Speir JA et al (2011) The 2.3-angstrom structure of porcine circovirus 2. J Virol 85:7856–7862. https://doi.org/10.1128/JVI.05863-11
Smith GP (1985) Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315–1317. https://doi.org/10.1126/science.4001944
Omidfar K, Daneshpour M (2015) Advances in phage display technology for drug discovery. Expert Opin Drug Discov 10:651–669. https://doi.org/10.1517/17460441.2015.1037738
Santos PS, Nascimento R, Rodrigues LP et al (2012) Functional epitope core motif of the Anaplasma marginale major surface protein 1a and its incorporation onto bioelectrodes for antibody detection. PLoS One. https://doi.org/10.1371/journal.pone.0033045
Wang M, Zhai L, Yu W et al (2018) Identification of a protective B-cell epitope of the Staphylococcus aureus GapC protein by screening a phage-displayed random peptide library. PLoS One 13:1–17. https://doi.org/10.1371/journal.pone.0190452
Wen X, Sun J, Wang X et al (2015) Identification of a novel linear epitope on the NS1 protein of avian influenza virus. BMC Microbiol 15:1–9. https://doi.org/10.1186/s12866-015-0507-4
Yang WJ, Lai JF, Peng KC et al (2005) Epitope mapping of Mycoplasma hyopneumoniae using phage displayed peptide libraries and the immune responses of the selected phagotopes. J Immunol Methods 304:15–29. https://doi.org/10.1016/j.jim.2005.05.009
Mahé D, Blanchard P, Truong C et al (2000) Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J Gen Virol 81:1815–1824. https://doi.org/10.1099/0022-1317-81-7-1815
Lekcharoensuk P, Morozov I, Paul PS, et al (2004) Epitope mapping of the major capsid protein of type 2 porcine circovirus ( PCV2 ) by using chimeric PCV1 and PCV2 epitope mapping of the major capsid protein of type 2 porcine circovirus ( PCV2 ) by using chimeric PCV1 and PCV2. 78:8135–8145. https://doi.org/10.1128/JVI.78.15.8135
Bin Shang S, Jin YL, Jiang XT et al (2009) Fine mapping of antigenic epitopes on capsid proteins of porcine circovirus, and antigenic phenotype of porcine circovirus Type 2. Mol Immunol 46:327–334. https://doi.org/10.1016/j.molimm.2008.10.028
Ge M, Yan A, Luo W et al (2013) Epitope screening of the PCV2 Cap protein by use of a random peptide-displayed library and polyclonal antibody. Virus Res 177:103–107. https://doi.org/10.1016/j.virusres.2013.06.018
Salgado RL, Vidigal PMP, Gonzaga NF et al (2015) A porcine circovirus-2 mutant isolated in Brazil contains low-frequency substitutions in regions of immunoprotective epitopes in the capsid protein. Arch Virol 160:2741–2748. https://doi.org/10.1007/s00705-015-2567-z
Carneiro AP, Reis CF, Morari EC et al (2014) A putative OTU domain-containing protein 1 deubiquitinating enzyme is differentially expressed in thyroid cancer and identifies less-aggressive tumours. Br J Cancer 111:1–8. https://doi.org/10.1038/bjc.2014.331
Barbas CF III, Burton DR, Scott JK, Silverman GJ (2001) Phage display: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview
McKinney MM, Parkinson A (1987) A simple, non-chromatographic procedure to purify immunoglobulins from serum and ascites fluid. J Immunol Methods 96:271–278. https://doi.org/10.1016/0022-1759(87)90324-3
Gerber PF, Galinari GCF, Silva MX et al (2009) Distribution of antibodies against porcine circovirus type-2 (PCV2) in single site and multi-site farrow-to-finish farms in Brazil. Res Vet Sci 87:488–491. https://doi.org/10.1016/j.rvsc.2009.04.013
Shanmugam A, Suriano R, Goswami N et al (2011) Identification of peptide mimotopes of gp96 using single-chain antibody library. Cell Stress Chaperones 16:225–234. https://doi.org/10.1007/s12192-010-0234-6
Horlen KP, Dritz SS, Nietfeld JC et al (2008) A field evaluation of mortality rate and growth performance in pigs vaccinated against porcine circovirus type 2. J Am Vet Med Assoc 232:906–912. https://doi.org/10.2460/javma.232.6.906
Martelli P, Ferrari L, Morganti M et al (2011) One dose of a porcine circovirus 2 subunit vaccine induces humoral and cell-mediated immunity and protects against porcine circovirus-associated disease under field conditions. Vet Microbiol 149:339–351. https://doi.org/10.1016/j.vetmic.2010.12.008
Fort M, Sibila M, Nofrarías M et al (2010) Porcine circovirus type 2 (PCV2) Cap and Rep proteins are involved in the development of cell-mediated immunity upon PCV2 infection. Vet Immunol Immunopathol 137:226–234. https://doi.org/10.1016/j.vetimm.2010.05.013
Steiner E, Balmelli C, Gerber H et al (2009) Cellular adaptive immune response against porcine circovirus type 2 in subclinically infected pigs. BMC Vet Res 5:45. https://doi.org/10.1186/1746-6148-5-45
Koinig HC, Talker SC, Stadler M et al (2015) PCV2 vaccination induces IFN-γ/TNF-α co-producing T cells with a potential role in protection. Vet Res 46:1–13. https://doi.org/10.1186/s13567-015-0157-4
Meng X-J (2013) Porcine Circovirus Type 2 (PCV2): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci 1:43–64. https://doi.org/10.1146/annurev-animal-031412-103720
Meerts P, Gucht SVAN, Cox E et al (2005) correlation between type of adaptive immune response against porcine circovirus type 2 and level of virus replication. Viral Immunol 18:333–341. https://doi.org/10.1089/vim.2005.18.333
Fort M, Olvera A, Sibila M (2007) Detection of neutralizing antibodies in postweaning multisystemic wasting syndrome ( PMWS ) -affected and non-PMWS-affected pigs. Vet Microbiol 125:244–255. https://doi.org/10.1016/j.vetmic.2007.06.004
Gamage LNA, Ellis J, Hayes S (2009) Immunogenicity of bacteriophage lambda particles displaying porcine Circovirus 2 (PCV2) capsid protein epitopes. Vaccine 27:6595–6604. https://doi.org/10.1016/j.vaccine.2009.08.019
Mosmann TR, Coffman RL (1989) Th1 AND Th2 CELLS: different patterns of lymphokine secretion lead to different functional properties. Ann Rev Immunol 7:145–173. https://doi.org/10.1146/annurev.iy.07.040189.001045
Lou Z, Li X, Li Z et al (2011) Expression and antigenicity characterization for truncated capsid protein of porcine circovirus type 2. Can J Vet Res 75:61–64
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
We thank the Brazilian Government Agencies. This research was funded by the Coordination for the Improvement of Higher Education Personnel - CAPES (grant number 23038.004678/2015-24), National Council for Scientific and Technological Development - CNPq (grant number 304727/2016-4), Foundation for Research Support of the State of Minas Gerais - FAPEMIG (grant numbers PPM-00796-15, CVZ-APQ-01327-14, CBB-RED-00005/14).
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
This project complied with the principles of the Commission for Ethics in Animal Experimentation of the Federal University of Viçosa (UFV) under protocol n°39/2012. All authors contributed to this work and agreed to its publication.
Additional information
Handling Editor: Sheela Ramamoorthy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2018_3816_MOESM1_ESM.jpg
Supplementary Fig 1S. Expression of scFv clones. 96-well plates were sensitized with the supernatant from the clones, incubated with anti-HA and developed with OPD solution.
705_2018_3816_MOESM2_ESM.jpg
Supplementary Fig 2S. Specificity of scFv fragments against Cap-PCV2 and complementarity determining regions (CDR). A) Binding capacity of scFv fragments to Cap-PCV2 and BSA determined by ELISA. B) Binding capacity of F1 and F5 scFv fragments to Cap-PCV2 assessed by Western blotting. Arrows indicate the stained Cap-PCV2 band (about 30 kDa). C+: Pig anti-PCV2 polyclonal antibodies; C-: negative control for primary antibody (PBS); PM: molecular weight standard. C) CDR sequences of the selected scFv fragments. *It was not possible to identify CDR3 sequences according to IgBlast database.
Rights and permissions
About this article
Cite this article
Santos, M.R., Assao, V.S., Santos, F.d.A. et al. Utilization of phage display to identify antigenic regions in the PCV2 capsid protein for the evaluation of serological responses in mice and pigs. Arch Virol 163, 1877–1887 (2018). https://doi.org/10.1007/s00705-018-3816-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-3816-8