Abstract
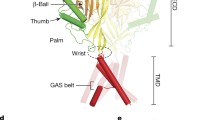
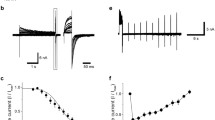
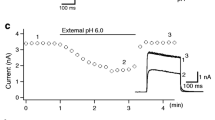
The bile acid-sensitive ion channel (BASIC) is a member of the ENaC/degenerin family of ion channels. It is activated by bile acids and inhibited by extracellular Ca2+. The aim of this study was to explore the molecular mechanisms mediating these effects. The modulation of BASIC function by extracellular Ca2+ and tauro-deoxycholic acid (t-DCA) was studied in Xenopus laevis oocytes heterologously expressing human BASIC using the two-electrode voltage-clamp and outside-out patch-clamp techniques. Substitution of aspartate D444 to alanine or cysteine in the degenerin region of BASIC, a region known to be critically involved in channel gating, resulted in a substantial reduction of BASIC Ca2+ sensitivity. Moreover, mutating D444 or the neighboring alanine (A443) to cysteine significantly reduced the t-DCA-mediated BASIC stimulation. A combined molecular docking/simulation approach demonstrated that t-DCA may temporarily form hydrogen bonds with several amino acid residues including D444 in the outer vestibule of the BASIC pore or in the inter-subunit space. By these interactions, t-DCA may stabilize the open state of the channel. Indeed, single-channel recordings provided evidence that t-DCA activates BASIC by stabilizing the open state of the channel, whereas extracellular Ca2+ inhibits BASIC by stabilizing its closed state. In conclusion, our results highlight the potential role of the degenerin region as a critical regulatory site involved in the functional interaction of Ca2+ and t-DCA with BASIC.











Similar content being viewed by others
References
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. https://doi.org/10.1093/bioinformatics/bti770
Baconguis I, Gouaux E (2012) Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature 489:400–405. https://doi.org/10.1038/nature11375
Baconguis I, Bohlen CJ, Goehring A, Julius D, Gouaux E (2014) X-ray structure of acid-sensing ion channel 1-snake toxin complex reveals open state of a Na+-selective channel. Cell 156:717–729. https://doi.org/10.1016/j.cell.2014.01.011
Berendsen H, Postma J, van Gunsteren W, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690. https://doi.org/10.1063/1.448118
Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. https://doi.org/10.1093/nar/gku340
Bussi G, Donadio D, Parrinello M (2007) Canonical sampling through velocity rescaling. J Chem Phys 126:014101. https://doi.org/10.1063/1.2408420
Case D, Babin V, Berryman J et al. (2014) Amber 14. University of California
Chen X, Minofar B, Jungwirth P, Allen HC (2010) Interfacial molecular organization at aqueous solution surfaces of atmospherically relevant dimethyl sulfoxide and methanesulfonic acid using sum frequency spectroscopy and molecular dynamics simulation. J Phys Chem B 114:15546–15553. https://doi.org/10.1021/jp1078339
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N · log (N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092. https://doi.org/10.1063/1.464397
Dawson RJ, Benz J, Stohler P, Tetaz T, Joseph C, Huber S, Schmid G, Hugin D, Pflimlin P, Trube G, Rudolph MG, Hennig M, Ruf A (2012) Structure of the acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1. Nat Commun 3:936. https://doi.org/10.1038/ncomms1917
Diakov A, Korbmacher C (2004) A novel pathway of ENaC activation involves an SGK1 consensus motif in the C-terminus of the channel’s α-subunit. J Biol Chem 279:38134–38142. https://doi.org/10.1074/jbc.M403260200
Diakov A, Bera K, Mokrushina M, Krueger B, Korbmacher C (2008) Cleavage in the γ-subunit of the epithelial sodium channel (ENaC) plays an important role in the proteolytic activation of near-silent channels. J Physiol 586:4587–4608. https://doi.org/10.1113/jphysiol.2008.154435
Diakov A, Nesterov V, Mokrushina M, Rauh R, Korbmacher C (2010) Protein kinase B alpha (PKBalpha) stimulates the epithelial sodium channel (ENaC) heterologously expressed in Xenopus laevis oocytes by two distinct mechanisms. Cell Physiol Biochem 26:913–924. https://doi.org/10.1159/000324000
Dickson CJ, Madej BD, Skjevik AA, Betz RM, Teigen K, Gould IR, Walker RC (2014) Lipid14: the amber lipid force field. J Chem Theory Comput 10:865–879. https://doi.org/10.1021/ct4010307
Fujimoto A, Kodani Y, Furukawa Y (2017) Modulation of the FMRFamide-gated Na+ channel by external Ca2+. Arch Eur J Physiol 469:1335–1347. https://doi.org/10.1007/s00424-017-2021-z
Gonzales EB, Kawate T, Gouaux E (2009) Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature 460:599–604. https://doi.org/10.1038/nature08218
Guex N, Peitsch MC, Schwede T (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30 Suppl 1:S162–S173. https://doi.org/10.1002/elps.200900140
Haerteis S, Krueger B, Korbmacher C, Rauh R (2009) The δ-subunit of the epithelial sodium channel (ENaC) enhances channel activity and alters proteolytic ENaC activation. J Biol Chem 284:29024–29040. https://doi.org/10.1074/jbc.M109.018945
Haerteis S, Krappitz A, Krappitz M, Murphy JE, Bertog M, Krueger B, Nacken R, Chung H, Hollenberg MD, Knecht W, Bunnett NW, Korbmacher C (2014) Proteolytic activation of the human epithelial sodium channel by trypsin IV and trypsin I involves distinct cleavage sites. J Biol Chem 289:19067–19078. https://doi.org/10.1074/jbc.M113.538470
Ilyaskin A, Diakov A, Sticht H, Korbmacher C, Haerteis S (2016) The degenerin region of the human bile acid sensitive ion channel is involved in channel inhibition by calcium and activation by bile acids. Acta Physiol 216(S707):70–84. https://doi.org/10.1111/apha.12671
Ilyaskin AV, Diakov A, Korbmacher C, Haerteis S (2016) Activation of the human epithelial sodium channel (ENaC) by bile acids involves the degenerin site. J Biol Chem 291:19835–19847. https://doi.org/10.1074/jbc.M116.726471
Ilyaskin AV, Diakov A, Korbmacher C, Haerteis S (2017) Bile acids potentiate proton-activated currents in Xenopus laevis oocytes expressing human acid-sensing ion channel (ASIC1a). Physiol Rep 5:e13132. https://doi.org/10.14814/phy2.13132
Irwin JJ, Shoichet BK (2005) ZINC-a free database of commercially available compounds for virtual screening. J Chem Inf Model 45:177–182. https://doi.org/10.1021/ci049714+
Jasti J, Furukawa H, Gonzales EB, Gouaux E (2007) Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature 449:316–323
de Jong DH, Singh G, Bennett WF, Arnarez C, Wassenaar TA, Schäfer LV, Periole X, Tieleman DP, Marrink SJ (2013) Improved parameters for the Martini coarse-grained protein force field. J Chem Theory Comput 9:687–697. https://doi.org/10.1021/ct300646g
Kellenberger S, Schild L (2015) International Union of Basic and Clinical Pharmacology. XCI. Structure, function, and pharmacology of acid-sensing ion channels and the epithelial Na+ channel. Pharmacol Rev 67:1–35. https://doi.org/10.1124/pr.114.009225
Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T (2009) The SWISS-MODEL repository and associated resources. Nucleic Acids Res 37:D387–D392. https://doi.org/10.1093/nar/gkn750
Korbmacher C, Volk T, Segal AS, Boulpaep EL, Frömter E (1995) A calcium-activated and nucleotide-sensitive nonselective cation channel in M-1 mouse cortical collecting duct cells. J Membr Biol 146:29–45
Lefèvre CM, Diakov A, Haerteis S, Korbmacher C, Gründer S, Wiemuth D (2014) Pharmacological and electrophysiological characterization of the human bile acid-sensitive ion channel (hBASIC). Arch Eur J Physiol 466:253–263. https://doi.org/10.1007/s00424-013-1310-4
Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78:1950–1958. https://doi.org/10.1002/prot.22711
Madej BD, Gould IR, Walker RC (2015) A parameterization of cholesterol for mixed lipid bilayer simulation within the Amber Lipid14 force field. J Phys Chem B 119:12424–12435. https://doi.org/10.1021/acs.jpcb.5b04924
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. https://doi.org/10.1002/jcc.21256
Páll S, Hess B (2013) A flexible algorithm for calculating pair interactions on SIMD architectures. Comput Phys Commun 184:2641–2650. https://doi.org/10.1016/j.cpc.2013.06.003
Páll S, Abraham MJ, Kutzner C, Hess B, Lindahl E (2014) Tackling exascale software challenges in molecular dynamics simulations with GROMACS. In: Markidis S., Laure E. (eds) Solving Software Challenges for Exascale. EASC 2014. Lecture Notes in Computer Science, V. 8759. Springer, Cham
Paukert M, Babini E, Pusch M, Gründer S (2004) Identification of the Ca2+ blocking site of acid-sensing ion channel (ASIC) 1: implications for channel gating. J Gen Physiol 124:383–394. https://doi.org/10.1085/jgp.200308973
Pluhackova K, Kirsch SA, Han J, Sun L, Jiang Z, Unruh T, Böckmann RA (2016) A critical comparison of biomembrane force fields: structure and dynamics of model DMPC, POPC, and POPE bilayers. J Phys Chem B 120:3888–3903. https://doi.org/10.1021/acs.jpcb.6b01870
Rauh R, Diakov A, Tzschoppe A, Korbmacher J, Azad AK, Cuppens H, Cassiman JJ, Dotsch J, Sticht H, Korbmacher C (2010) A mutation of the epithelial sodium channel associated with atypical cystic fibrosis increases channel open probability and reduces Na+ self inhibition. J Physiol 588:1211–1225. https://doi.org/10.1113/jphysiol.2009.180224
Sakai H, Lingueglia E, Champigny G, Mattei MG, Lazdunski M (1999) Cloning and functional expression of a novel degenerin-like Na+ channel gene in mammals. J Physiol 519(Pt 2):323–333
Sanner MF (1999) Python: a programming language for software integration and development. J Mol Graph Model 17:57–61
Schaefer L, Sakai H, Mattei M, Lazdunski M, Lingueglia E (2000) Molecular cloning, functional expression and chromosomal localization of an amiloride-sensitive Na+ channel from human small intestine. FEBS Lett 471:205–210
Schmidt A, Lenzig P, Oslender-Bujotzek A, Kusch J, Lucas SD, Gründer S, Wiemuth D (2014) The bile acid-sensitive ion channel (BASIC) is activated by alterations of its membrane environment. PLoS One 9:e111549. https://doi.org/10.1371/journal.pone.0111549
Schmidt A, Lohrer D, Alsop RJ, Lenzig P, Oslender-Bujotzek A, Wirtz M, Rheinstadter MC, Gründer S, Wiemuth D (2016) A cytosolic amphiphilic α-helix controls the activity of the bile acid-sensitive ion channel (BASIC). J Biol Chem 291:24551–24565. https://doi.org/10.1074/jbc.M116.756437
Siu SW, Pluhackova K, Böckmann RA (2012) Optimization of the OPLS-AA force field for long hydrocarbons. J Chem Theory Comput 8:1459–1470. https://doi.org/10.1021/ct200908r
Sun Y, Kollman PA (1995) Hydrophobic solvation of methane and nonbond parameters of the TIP3P water model. J Comput Chem 16:1164–1169. https://doi.org/10.1002/jcc.540160910
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334
Wassenaar TA, Pluhackova K, Böckmann RA, Marrink SJ, Tieleman DP (2014) Going backward: a flexible geometric approach to reverse transformation from coarse grained to atomistic models. J Chem Theory Comput 10:676–690. https://doi.org/10.1021/ct400617g
Wassenaar TA, Ingólfsson HI, Böckmann RA, Tieleman DP, Marrink SJ (2015) Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. J Chem Theory Comput 11:2144–2155. https://doi.org/10.1021/acs.jctc.5b00209
Wiemuth D, Gründer S (2011) The pharmacological profile of brain liver intestine Na+ channel: inhibition by diarylamidines and activation by fenamates. Mol Pharmacol 80:911–919. https://doi.org/10.1124/mol.111.073726
Wiemuth D, Sahin H, Falkenburger BH, Lefèvre CM, Wasmuth HE, Gründer S (2012) BASIC-a bile acid-sensitive ion channel highly expressed in bile ducts. FASEB J 26:4122–4130. https://doi.org/10.1096/fj.12-207043
Wiemuth D, Sahin H, Lefèvre CM, Wasmuth HE, Gründer S (2013) Strong activation of bile acid-sensitive ion channel (BASIC) by ursodeoxycholic acid. Channels (Austin) 7:38–42. https://doi.org/10.4161/chan.22406
Wiemuth D, Lefèvre CM, Heidtmann H, Gründer S (2014) Bile acids increase the activity of the epithelial Na+ channel. Arch Eur J Physiol 466:1725–1733. https://doi.org/10.1007/s00424-013-1403-0
Yesylevskyy SO, Schäfer LV, Sengupta D, Marrink SJ (2010) Polarizable water model for the coarse-grained MARTINI force field. PLoS Comput Biol 6:e1000810. https://doi.org/10.1371/journal.pcbi.1000810
Acknowledgements
The expert technical assistance of Ralf Rinke is gratefully acknowledged. This work was supported by grants of the Deutsche Forschungsgemeinschaft (DFG) (HA 6655/1-1 to S.H.), the DFG Research Training Group 1962/1, Dynamic Interactions at Biological Membranes—From Single Molecules to Tissue (S.A.K. and R.A.B.), and the Johannes and Frieda Marohn Stiftung (C.K.). Part of this work has been published in abstract form [20]. We thank Kristyna Pluhackova for support in the parameterization procedure.
Abbreviations
BASIC Bile acid-sensitive ion channel
ENaC Epithelial sodium channel
ASIC1 Acid-sensing ion channel 1
P o Open probability
t-DCA Tauro-deoxycholic acid
TMD, transmembrane domain
Author information
Authors and Affiliations
Contributions
Alexandr V. Ilyaskin, Alexei Diakov, and Sonja A. Kirsch performed the experiments, analyzed the data, and prepared the figures (Alexandr V. Ilyaskin, Alexei Diakov: electrophysiological experiments; Alexandr V. Ilyaskin: molecular docking simulations; Sonja A. Kirsch: molecular dynamics simulations). Alexandr V. Ilyaskin, Sonja A. Kirsch, Rainer A. Böckmann, Heinrich Sticht, Christoph Korbmacher, Silke Haerteis, and Alexei Diakov designed the study, interpreted the data, and wrote the paper. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(DOCX 1688 kb)
Rights and permissions
About this article
Cite this article
Ilyaskin, A.V., Kirsch, S.A., Böckmann, R.A. et al. The degenerin region of the human bile acid-sensitive ion channel (BASIC) is involved in channel inhibition by calcium and activation by bile acids. Pflugers Arch - Eur J Physiol 470, 1087–1102 (2018). https://doi.org/10.1007/s00424-018-2142-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00424-018-2142-z